Abstract
Background
Despite significant progress in eliminating malaria from the state of Odisha, India, the disease is still considered endemic. Artesunate plus sulfadoxine-pyrimethamine (AS + SP) has been introduced since 2010 as first-line treatment for uncomplicated Plasmodium falciparum malaria. This study aimed to investigate the prevalence of mutations associated with resistance to chloroquine (CQ), sulfadoxine-pyrimethamine (SP), and artesunate (ART) in P. falciparum parasites circulating in the state.
Methods
A total of 239 isolates of P. falciparum mono infection were collected during July 2018-November 2020 from the four different geographical regions of the state. Genomic DNA was extracted from 200 µL of venous blood and amplified using nested polymerase chain reaction. Mutations on gene associated with CQ (Pfcrt and Pfmdr1) were assessed by PCR amplification and restriction fragment length polymorphism, artemisinin (Pfk13) gene by DNA sequencing and SP (Pfdhfr and Pfdhps) genes by allele-specific polymerase chain reaction (AsPCR).
Results
The point mutation in Pfcrt (K76T) was detected 2.1%, in Pfmdr1 (N86Y) 3.4%, and no mutations were found in Pfkelch13 propeller domain. Prevalence of Pfdhfr, Pfdhps and Pfhdfr-Pfdhps (two locus) gene mutations were 50.43%, 47.05% and 49.79% respectively. The single, double, triple and quadruple point mutations in Pfdhfr gene was 11.2%, 8.2%, 17.2% and 3.4% while, in Pfdhps gene was 10.9%,19.5%, 9.5% and 2.7% respectively. Of the total 13 haplotypes found in Pfdhfr, 8 were detected for the first time in the state and of the total 26 haplotypes found in Pfdhps, 7 were detected for the fisrt time in the state. The linked quintuple mutation Pfdhfr (N51I-C59R-S108N)-Pfdhps (A437G-K540E) responsible for clinical failure (RIII level of resistance) of SP resistance and A16V-S108T mutation in Pfdhfr responsible for cycloguanil was absent.
Conclusion
The study has demonstrated a low prevalence of CQ resistance alleles in the study area. Despite the absence of the Pfkelch13 mutations, high prevalence of Pfdhfr and Pfdhps point mutations undermine the efficacy of SP partner drug, thereby threatening the P. falciparum malaria treatment policy. Therefore, continuous molecular and in vivo monitoring of ACT efficacy is warranted in Odisha.
Similar content being viewed by others
Background
Malaria caused by Plasmodium species occurs mainly in poor tropical and sub-tropical regions of the world. Of the five species causing human malaria, Plasmodium falciparum is the most lethal and accounts for more than 90% of the world's malaria deaths [1]. Although malaria mortality has been reduced by over a quarter around the world, its transmission still occurs in 99 countries. In 2020, an estimated 241 million malaria cases and 627,000 malaria deaths have occurred worldwide [2]. In Southeast Asia, India alone accounts for 85.2% of malaria cases and 2% of global malaria deaths [3]. While in India, the state of Odisha, with 4% of the total population of the country, accounted for 32.4% of the total malaria cases, 32.02% of P. falciparum cases, and 9.7% of malaria deaths in 2020 (NVBDCP, Govt. of India).
In absence of an effective vaccine, chemotherapy and chemoprophylaxis remain the principal means to minimize the mortality and morbidity burden due to malaria. As in other malaria-endemic countries of the world, chloroquine (CQ) was used in the national programme in India since 1961 as the first-line treatment for uncomplicated malaria for a prolonged period because of its safety profile and cost-effectiveness. After sustained use, the resistance of P. falciparum to CQ emerged first time in India in 1973 in the Karbi-Anglong district of Assam that subsequently spread across the country [4, 5]. Accordingly, the Indian drug policy was changed and the sulfadoxine-pyrimethamine (SP) combination was introduced in 1995 as a second-line treatment [6]. However, in 2004 the World Health Organization (WHO) technical advisory group recommended the use of combination anti-malarial therapy, particularly with artemisinin derivatives, in member countries for treating P. falciparum to delay the emergence of drug resistance. Consequently, artemisinin-based combination therapy (ACT) i.e. using artesunate + sulfadoxine-pyrimethamine (AS-SP) was first introduced in 2004 in CQ resistant areas and then implemented in the rest of the country in 2010 as the first-line treatment of P. falciparum malaria [7].
The effective implementation of any drug policy needs continuous monitoring of drug-resistant parasites in the field to determine the spread of resistance over wide areas. Since the identification of drug-resistant P. falciparum strains by in vitro assay and standard 28-day in vivo efficacy study are cumbersome, molecular markers have been proposed as an alternative tool to monitor resistance [8]. The point mutation in P. falciparum CQ transporter (Pfcrt) gene (K76T) and P. falciparum multidrug-resistance1 (Pfmdr1) gene (N86Y) have been found to be associated with CQ resistance [9, 10]. Resistance to SP drug combination has been shown to occur due to the point mutations in the P. falciparum dihydrofolate reductase (Pfdhfr) gene (A16V, C50R, N51I, C59R, S108T/N, and I164L) and P. falciparum dihydropteroate synthase (Pfdhps) gene (S436A/F, A437G, K540E, A581G, and A613S/T) [11]. Multiple mutation combinations of both Pfdhps and Pfdhfr were responsible in varying the level of SP resistance. While, mutations in the propeller domain of the Kelch-13 protein encoded by the P. falciparum Pfk13 gene have been associated with delayed parasite clearance due to resistance to artemisinin (ART) in the Greater Mekong Sub-regions of Southeast Asia and sub-Saharan regions of Africa [12,13,14,15].
India has pledged to eliminate malaria in the entire country by 2030 [16]. To achieve the target, wide coverage of molecular data on anti-malarial drug resistance is essential for proper implementation of drug treatment policy. Hence, the present study was undertaken to assess the prevalence of Pfcrt K76T and Pfmdr1 N86Y (responsible for chloroquine resistance), mutations of Pfdhfr and Pfdhps genes responsible for SP drug resistance and P. falciparum Pfk13 polymorphism associated with artemisinin (ART) treatment failure on P. falciparum isolates in Odisha, between 2018 and 2020, after ten years of the introduction of new drug policy. The data from the study could contribute to baseline information on the distribution of anti-malarial drug resistance, particularly in Odisha prior to malaria elimination.
Methods
Study setting and sample collection
This study was conducted between July 2018 to November 2020 among the patients attending government health facilities in different districts representing four geographical regions of the state (Eastern Ghat: Raygada, Kalahandi, Nuapada, Kandhamal, Gajapati, Northern Plateau: Mayurbhanj, Sundergarh, and Keonjhar, Central Tableland: Bargarh, Angul, Deogarh and, Coastal Belt: Cuttack, Khorda, and Ganjam) as shown in Fig. 1. Based on the overall annual parasitic index (API) of the districts as reported by NVBDCP, Odisha, 2016, the Eastern Ghat and Northern Plateau can be categorized as highly endemic (API > 10), the Central Tableland (API 5–10) as moderately endemic and Coastal Belt districts are very low endemic (API < 0.5). As per Indian drug policy, ACT has been used as a treatment for P. falciparum infection, and chloroquine + primaquine was used for P. vivax infection. As per the available literature the prevalence of CQ and SP drug-resistant haplotypes was high in the districts of Northern plateau, Eastern Ghat, moderate in the districts of Central Tableland and low in the districts of Coastal Belt [17, 18]. Suspected malaria cases were screened by the WHO evaluated Pf PAN Ag Rapid Diagnostic Test Kit (RDT) (SD-Biosensor, India) using finger-prick blood [19]. Approximately one mL of venous blood was collected in BD Vacutainer®EDTA vial from individuals found to be positive for malaria. The blood samples collected in the field were preserved at 40C at the local hospital and transported to the Indian Council of Medical Research (ICMR)-Regional Medical Research Centre (RMRC) Bhubaneswar laboratory within 24 h, maintaining a cold chain for further molecular analysis.
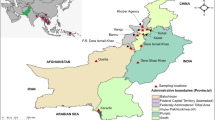
Map of India A and district of Odisha B indicating the study districts of four geographical regions. The base maps were taken from the website: https://www.burningcompass.com/countries/India/odisha-on-India-map-hd.html and was modified using Microsoft word and paint. The studied fourteen districts of the four different geographical regions marked by four distinct marks (i) Northern Plateau (purple), (ii) Eastern Ghat (red), (iii) Central Tableland (green) and (iv) Coastal Belt (yellow)
Diagnosis and speciation by PCR
Parasite genomic DNA was extracted from 200 μL of EDTA blood samples using QIAamp Blood DNA mini kit (QIAGEN, Germany) according to the manufacturer's instructions and eluted in 50 μL TE buffer. Nested PCR (nPCR) was performed to confirm the diagnosis and identification of the species using the species specific primers targeting 18SrRNA and cycling parameters as described by Snounou et al. [20] in a thermal cycler (Agilent Sure Cycler 8800, USA). Briefly, the primary PCR was performed on a 25 μL reaction mixture that contained 0.2 U of Taq DNA polymerase (GCC Biotech, India), 0.2 mM each dNTP (HIMEDIA Laboratories, India), 0.4 mM each primer (GCC Biotech, India) and 2.0 mM MgCl2 (GCC Biotech, India). The reaction was allowed to proceed for 35 cycles after an initial denaturation at 940 for 1 min, annealing at 500 for 1 min, and extension at 720 for 1 min; final extension was at 720 for 10 min. The nested PCR reaction condition was the same as primary PCR except for the annealing temperature, 550. PCR products were visualized under UV light following the electrophoresis on 2.0% (w/v) ethidium bromide stained agarose gel, and images were captured using a gel documentation system (Alpha Imager, USA). Previously diagnosed Plasmodium species specific DNA was used as positive control and genomic DNA extracted from uninfected individuals was used as negative control.
Analysis of Pfcrt, Pfmdr1 & Pfk13 genes
Genotyping of the resistant markers Pfcrt K76T and Pfmdr1 N86Y was carried out by PCR-Restriction fragment length polymorphism (RFLP) using the genomic DNA isolated from all the enrolled samples (n = 239) followed by sequencing for Pfcrt and Pfmdr1 haplotype analysis. Briefly, PCR protocol and the primers (Table 1) as described elsewhere [17] amplified the 264 bp Pfcrt gene spanning the codon region from 72 to 76, 78, 97 and the 603 bp pfmdr1 gene with codons 86,130,184. The PCR products were digested with Type II restriction digestion enzyme Apo I for detection of the Pfcrt sensitive/resistant genotype and Afl III for Pfmdr1 sensitive/resistant genotype. The Apo I digests the 264 bp Pfcrt PCR product into 128 bp and 136 bp fragments in case of the CQ wild (sensitive) allele, but the mutant allele associated with CQ resistance remains undigested. Similarly, 603 bp of Pfmdr1 PCR product when treated with Afl III the mutant (resistant) allele digested into 353 bp and 253 bp fragments, while the CQ/multidrug-sensitive (wild) genotypes remain undigested. The Pfkelch13 gene fragment was amplified by nested PCR protocols reported previously with modifications [12].
In case of Pfcrt, 212 nucleotide sequence fragments encompassing the K76T mutations responsible for CQ resistance, while 526 nucleotide sequence fragments of Pfmdr1 containing the N86Y mutations responsible for multidrug resistance and 793 nucleotide sequence fragment of Pfk13 gene containingN458Y, Y493H R539T, I543T and C580Yknown point mutation responsible of ART resistance were sequenced [17]. The sequences of Pfcrt, Pfmdr1, and Pfk13 found in the study have been deposited in Gene Bank via Bankit http://www.ncbi.nlm.nih.gov/Bankit (Accession # MZ678763-MZ678766 for Pfcrt, MZ054306, MZ054305, and MZ678767-MZ678769 for Pfmdr1, and MZ151068-MZ151071 for Pfk13 gene).
DNA sequence analysis
The DNA sequences were aligned; and population genetic parameters were calculated for each gene separately. Manual editing and alignment of DNA sequences was conducted using SeqMan, EditSeq, and MegAlign modules of the Laser gene computer program [17]. All the parameters were calculated using the computer program DNA Sequence polymorphism v6.12.03 (DnaSP) [21].
Analysis of Pfdhfr &Pfdhps genes
Allele specific polymerase chain reaction (AsPCR) assay was performed to investigate the presence of point mutations in Pfdhfr and Pfdhps gene associated with antifolate resistance as per the published protocol [22].
Ethics and consent
The Institutional Human Ethics Committee of the ICMR-RMRC Bhubaneswar has approved the study. Before the enrolment, the purpose of the study was explained to the participants in the local language, and verbal consent was obtained for blood sample collection and testing. One consent from the adult patient (above18 year age) and consent for children less than 18-year age from their parents or head of the house hold members in case of no parents, as per ICMR guidelines; written informed consent was obtained from patients or the parents/guardians of children prior to blood collection.
Data analysis
The data obtained was analysed using Microsoft Excel. Statistical analysis was carried out using P-values from a chi-square test for proportions using P-value of 0.05, comparing the relationship between different individual mutations, haplotypes, with respect to different geographical regions. The analysis was done using SPSS 20.0 (IBMCorp.2020, Chicago, IL).
Results
Of the total 557 RDT malaria positive samples, 476 were found to be positive for malaria by nested PCR. Amongst the PCR-positive samples, 239 were P. falciparum mono infection, 123 were non-falciparum malaria (NFM) (Plasmodium malariae: 7, Plasmodium ovale: 2 and Plasmodium vivax: 114) and 114 were P. falciparum mixed with NFM infections. The flow diagram of the selection of P. falciparum mono-infection samples used in the present study for molecular analysis of drug resistance genes has been depicted in Fig. 2 and the baseline characteristics of the enrolled cases has been shown in Table 2.
Analysis of Pfcrt, Pfmdr1 and Pfk13 gene
PCR–RFLP analysis was performed in 239 P. falciparum isolates have shown Pfcrt K76T point mutation in five isolates (2.1%), Pfmdr1N86Y point mutation in 8 isolates (3.4%) and no isolate carried Pfcrt K76T + Pfmdr1N86Y point mutations.
Single nucleotide mutations identified through DNA sequencing translated to amino acid substitutions in a subset of samples revealed two different types of haplotypes (CVIET and CVMNT) in isolates having Pfcrt 72-76 point mutation, the primary determinant of chloroquine (CQ) resistance, while analysis of wild type (Pfcrt K76) samples shown CVMNK haplotype. Of the two different kinds of haplotypes detected in mutant samples during the survey, both the haplotypes (CVIET and CVMNT) were found in the P. falciparum isolates collected from the Eastern Ghat (Raygada and Kandhamal), while the CVIET was found in Sundargarh district of Northern Plateau region and CVMNT in Bargarh district of Central Tableland region as shown in Table 3. SVMNT haplotype was not reported in the recent study. Similarly, sequencing of the Pfmdr1 mutant isolates detected by PCR–RFLP showed N86Y and N86Y/Y184F point mutations. There was no significant difference (χ2 = 6; P > 0.05) in the distribution of Pfcrt and Pfmdr1 associated with chloroquine drug resistant genes in the four different geographical regions of the state as shown in Table 3.
Though none of the validated or established mutations associated with artemisinin resistance detected in P. falciparum isolates subjected for DNA sequencing, six synonymous mutations that were not coding any proteins but changes only in change of nucleotides i.e. A160C, A208G, C210G, T211G, T212G, A251G were reported in this study, indicating the absence of P. falciparum genotype (Pfk13) associated with resistance to artemisinin in Odisha at present. The population genetic parameters for all the three genes responsible for anti-malarial resistance are displayed in Table 4. While the haplotype diversity was almost similar in all the three genes, the nucleotide diversities, as measured independently by theta (ϴ) and Pi (π), were variable across the three genes. Whereas relatively higher nucleotide diversities were found in the Pfcrt gene for both the parameters theta (ϴ) and Pi (π), the values were found to be lower in the Pfmdr1 and Pfk13 genes. The test of neutrality as measured by Tajima D, Fu and Li’s D, and Fu and Li’s F tests were not statistically significantly deviated from the model of neutral expectation in any of the three genes.
Analysis of Pfdhfr and Pfdhps genes
A total of 239 P. falciparum infected blood samples were analysed for mutations in six codons of the Pfdhfr gene (A16V, C50R, N51I, C59R, S108N/T and I164L) and five codons of the Pfdhps gene (S436F/A, A437G, K540E, A581G and A613S/T) to assess the level of antifolate drug pressure. Out of 239 samples, 232 were PCR positive for Pfdhfr and 221 for Pfdhps, while PCR could detect both Pfdhfr and Pfdhps genes in 119 (49.7%) of the samples. The Pfdhfr C59R mutation was found to be most prevalent (N = 97, 41.8%), followed by the C50R mutation (N = 93, 40.8%) and S108N mutation (N = 91, 39.2%), No isolate had the S108T mutation, while the N51I, I164L, and A16V mutations were found in 17.2% (N = 40), and 3.4% (N = 8) and 9.05% (N = 21) of the isolates respectively as shown in Table 5. There was no significant difference between χ2 = 8, P-value = 0.238 for A16V, N51I and I164L and, χ2 = 12, P-value = 0.213 for C50R, C59R and S108N individual amino acid mutation of the Pfdhfr gene in the four different geographical regions of the state as shown in Table 5. The wild-type Pfdhfr sequence (ACNCSI) at all six codons was prevalent in 49.6% (N = 115) of the isolates. Amongst the total isolates 26 (11.2%) had a single mutation, 19 (8.2%) had double, 40 (17.2%) had triple, 23 (9.9%) had quadruple, 8 (3.4%) had quintuple and 1 (0.43%) had sextuple mutation. The most frequent triple mutation sequence was ARNRS/NI (N = 18, 7.8%) ARIRS/SI (N = 17, 7.3%) and the quadruple mutation sequence was ARIRS/NI (N = 12, 5.2%) VRNRS/NI (N = 11, 4.7%). In our sample (N = 232) total 13 different haplotypes have been observed in the Pfdhfr gene in four different geographical regions of the state as shown in Table 6. There was no significant difference (as P-value > 0.05) in 13 haplotypes of Pfdhfr gene with respect to four different geographic regions of the state i.e. Northern Plateau; χ2 = 78, P-value = 0.294, Eastern Ghat; χ2 = 117, P-value = 0.261, Central Tableland; χ2 = 39, P-value = 0.336, and, Coastal Belt; χ2 = 26, P-value = 0.353, as shown in Table 7.
Of the 221 samples PCR positive for Pfdhps, 117 (52.9%) had the wild-type sequences (SAKAA) at all five codons. The maximum number of mutations were found at codon S436A (N = 59, 26.7%), followed by A613S (N = 39, 17.6%), K540E (N = 38, 17.2%), A581G (N = 31, 14.0%), S436F (N = 28, 12.7%), A437G (N = 26, 11.8%) and A613T (N = 23, 10.4%) as shown in Table 5. There was no significant difference (as P-value > 0.05) between S436F, A437G, A613S, A581G, A613T (χ2 = 12, P-value = 0.213) and for S436A, K540E (χ2 = 8, P-value = 0.238) of the Pfdhfr gene codons in the four different geographical regions of the state as shown in Table 5. In comparison to single (N = 24, 10.9%), triple (N = 21, 9.5%), quadruple (N = 6, 2.7%), quintuple (N = 6, 2.7%), and sextuple (N = 4, 1.8%) mutations, double mutations in Pfdhps gene had more prevalent (N = 43, 19.5%) as shown in Table 6. Amongst the double mutations, the sequences with S/AAKAS/A (N = 15, 6.8%) and S/AAEAA/A (N = 13, 5.9%), and amongst the triple mutations the sequences with S/AAKGA/A (N = 8, 3.6%), S/AAEAS/A (N = 7, 3.2%), F/AGKAS/A (N = 7, 3.2%), F/SGEGA/T (N = 5, 2.3%) were common, as shown in Table 6. There was no significant difference (as P-value > 0.05) in 26 haplotypes of Pfdhps gene with respect to four different geographic regions of the state i.e. Northern Plateau; χ2 = 130, P-value = 0.326, Eastern Ghat; χ2 = 182, P-value = 0.343, Central Tableland and Coastal Belt; χ2 = 78, P-value = 0.384, as shown in Table 7.
Plasmodium falciparum dhfr-dhps two-locus mutation analysis
The P. falciparum Pfdhfr-Pfdhps two-locus mutation analysis carried out in 119 (49.79%) isolates have revealed 3 different genotypes in Coastal Belt, 5 in Central Tableland, 29 in Northern Plateau and ≥ 40 in Eastern Ghat regions. However, no isolate with Pfdhfr triple (N51I/C59R/S108N) mutation in combination with Pfdhps double (A437G/K540E) mutation, a useful predictor of SP treatment failure, was found in the studied sample.
Discussion
The present study conducted during 2018–2020 has demonstrated a low prevalence (2.1%) of Pfcrt K76T mutation associated with resistance to CQ in P. falciparum isolates circulating in Odisha. Moreover, the same low percentage of mutation has also been detected for Pfmdr1 N86Y (3.4%). In contrast, a high prevalence of Pfcrt K76T (67.5%) and Pfmdr1 N86Y (80%) was observed in another study conducted before CQ withdrawal from the state during 2008–2010 [17]. The result obtained is similar to the observations made in Malawi, Tanzania, Mozambique, Northern Uganda, Saudi Arabia [23,24,25,26,27], but in contrast to southern Benin [28] and in other parts of India reported recently [29,30,31,32]. The present study, although limited to a small number of samples, indicates not only the presence of three types haplotypes (CVMNK: wild type, CVIET mutant type: believed to be of Southeast Asian origin, CVMNT mutant type: believed to be of African Origin) but also inform the high genetic diversity present in field isolates of P. falciparum for CQ drug-resistant genes in Odisha, India. Interestingly, the wild type CVMNK haplotype of the Pfcrt gene was found in 76.47% of the isolates, which is in contrast to findings from other studies in Indian P. falciparum as documented in earlier studies [33]. It is also argued that Odisha might have served as the epicentre for the distribution of chloroquine-resistant P. falciparum parasites to other parts of India [34, 35].
More than 200 non-synonymous mutations have been identified in K13 protein from P. falciparum strains in different malaria endemic countries and 50 of them are shown to be associated with ART treatment failure [36]. Studies conducted in India have identified fourteen K13 mutations in K189T, F446I, A481V, G533A/S, R539T, S549Y, R561H, A578S, M579T, G625R, N657H, N672S, A675V and D702N [37,38,39] and two of them (R539T, G625R) are shown to be associated with ART resistance [37]. No non-synonymous mutations have been observed during the present study despite occurrence of silent/synonymous mutations. DNA sequence polymorphism study that not only informs distribution of different haplotypes, but also the evolutionary potentiality of mutation in the drug-resistant genes that can directly translate to molecular epidemiology of human diseases like malaria. Several studies employing this methodology on the three genes of interest for malaria public health have been conducted worldwide, which has immensely helped in determining intervention through therapeutic measures and change in drug policy in different countries [40,41,42,43,44].
Sequential accumulation of S436F/A, A437G, K540E, A581G, A613S/T mutations in Pfdhps [45]and A16V, C50R, N51I, C59R, and S108T/N mutations in Pfdhfr [46] leads to the development of resistance to sulphadoxine and pyrimethamine respectively in P. falciparum isolates [47]. The primary mutation being A437G/ K540E in Pfdhps and S108N/ C59R in Pfdhfr as per the findings from different malaria endemic regions of the world including India [22, 48,49,50].The high proportion of mutation at codon C59R (41.4%), C50R (38.4%) and S108N (32.8%) mutations in the Pfdhfr gene than at codon S436A (26.7%),A613S (17.6%) and K540E (17.3%) mutations in Pfdhps gene indicate that these are the key point mutations and further the overall low prevalence of point mutations in Pfdhps (47.05%) gene sequence compared to Pfdhfr (50.4%) confirms that the mutations associated with parasite resistance to SP appeared earlier on the Pfdhfr than those affecting the Pfdhps [28]. The prevalence of single, double, triple, quadruple or quintuple mutation in Pfdhfr and Pfdhps observed in this study reflects the current level of sensitivity of P. falciparum to SP. Moreover, mutation at codon C59 and S108 along with codons A16, C50 and N51 in Pfdhfr (~ 38%) and codon A437 and K540 along with codons S436, A581 and A613 in Pfdhps (~ 24%) during the present study strongly predicts the decreasing treatment response as reported earlier in Western and Central Africa [51,52,53]. Out of the 12 Pfdhfr mutant genotypes found in the P. falciparum isolates in the state, 3 mutant genotypes (ACNCS/NI, ACNRS/SI, ARNCS/NI) have been reported earlier in India including Odisha [22], while 8 mutant genotypes (ARNRS/SI ARNRS/NI ACNRS/SL ARIRS/SI ARNRS/NL, ARIRS/NI, VRIRS/NI, VRIRS/NL) have been found for the first time in the state. Similar is the situation in the case of Pfdhps gene, in which S/AAKAS/A, S/AAEAS/A, F/AGKAS/A, F/SGEGA/T, F/SGEGS/T, F/AGEGA/T and F/AGE/GAA genotypes have been detected for the first time in the state in addition to S/AAEAA/A and S/SAKGA/T genotypes reported from India including Odisha. Prevalence of such unique multiple mutations in Pfdhfr as well as Pfdhps in the state indicates emergence of resistance to SP, the currently used partner drug of ACT in the state, as observed earlier in Kenya, Thailand and Vietnam [22]. But, absence of linked N51I-C59R-S108N codons in Pfdhfr and A437G-K540Ecodons in Pfdhps indicates that P. falciparum isolates circulating in this part of the country have not developed RIII (highest) level of resistance [54]. Similarly, absence of A16V-S108T mutation in Pfdhfr responsible for cycloguanil resistance in the present study might be because cycloguanil-proguanil has not yet been introduced for the treatment of malaria in India [7].
Limitation of the study
There are some limitations that should be considered when interpreting the findings of the present study. First, the total number of the collected P. falciparum isolates was small and disproportionate to different geographical region. Second, the molecular analysis (DNA sequencing) has been done in a subset of samples instead of total sample. Third, the copy number of Pfmdr1 has not been analysed.
Conclusion
This was the first molecular study carried out in the state of Odisha (India), after a gap of ten years of CQ withdrawal, focusing on mutations of Pfcrt and Pfmdr1 genes strongly associated with CQ, Pfkelch13 associated with ART, and Pfdhps and Pfdhfr genes strongly associated with resistance to SP, the partner drug used with ACT in the current drug policy. This study showed low prevalence of resistance to marker CQ that dramatically contrasted with our earlier study in the state. This study found an absence of Pfkelch13 mutations associated with ART resistance in P. falciparum isolates. However, the prevalence of triple, quadruple, quintuple and sextuple mutated Pfdhfr-Pfdhps genotypes sounds an alarm and, therefore, continuous molecular and in vivo monitoring of ACT is recommended for ensuring proper malaria control.
Availability of data and materials
All data generated or analysed during this study are included in the article. The nucleotide sequences, except Pfdhfr and Pfdhps, generated have been deposited in Gene Bank via Bankit http://www.ncbi.nlm.nih.gov/Bankit/ and are available in the database under the accession numbers as indicated in the text.
References
World malaria report. Geneva: World Health Organization; 2019
World malaria report. Geneva: World Health Organization; 2021
WHO. World Malaria Report. 20 years of global progress and challenges. Geneva: World Health Organization; 2020. p. 2020.
Sehgal PN, Sharma MID, Sharma SL, Gogai S. Resistance to chloroquine in falciparum malaria in Assam state. India J Commun Dis. 1973;5:175–80.
Farooq U, Mahajan RC. Drug resistance in malaria. J Vector Borne Dis. 2004;41:45–53.
Anvikar AR, Arora U, Sonal GS, Mishra N, Shahi B, Savargaonkar D, et al. Antimalarial drug policy in India: past, present & future. Indian J Med Res. 2014;139:205–15.
National Institute of Malaria Research (NIMR). Guidelines for diagnosis and treatment of malaria in India, 3rd ed. New Delhi: NIMR; 2014.
Bloland PB. Drug resistance in malaria. Geneva: World Health Organization; 2001.
Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, Kümpornsin K, et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun. 2018;9:25–8.
Njiro BJ, Mutagonda RF, Chamani AT, Mwakyandile T, Sabas D, Bwire GM. Molecular surveillance of chloroquine-resistant Plasmodium falciparum in sub-Saharan African countries after withdrawal of chloroquine for treatment of uncomplicated malaria: a systematic review. J Infect Public Health. 2022;15:550–7.
Jiang T, Chen J, Fu H, Wu K, Yao Y, Urbano J, et al. High prevalence of Pfdhfr – Pfdhps quadruple mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates from Bioko Island Equatorial Guinea. Malar J. 2019;18:101.
Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5.
Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–67.
Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, et al. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–20.
Stokes BH, Ward KE, Fidock DA. Evidence of artemisinin-resistant malaria in Africa. N Engl J Med. 2022;386:1385–6.
Lal AA, Rajvanshi H, Jayswar H, Das A, Bharti P. Malaria elimination: using past and present experience to make malaria-free India by 2030. J Vector Borne Dis. 2019;56:60–5.
Das Sutar SK, Gupta B, Ranjit M, Kar SK, Das A. Sequence analysis of coding DNA fragments of pfcrt and pfmdr-1 genes in Plasmodium falciparum isolates from Odisha. India Mem Inst Oswaldo Cruz. 2011;106:78–84.
Kar NP, Chauhan K, Nanda N, Kumar A, Carlton JM, Das A. Comparative assessment on the prevalence of mutations in the Plasmodium falciparum drug-resistant genes in two different ecotypes of Odisha state. India Infect Genet Evol. 2016;41:47–55.
Malaria rapid diagnostic test performance: results of WHO product testing of malaria RDTs: round 8 (2016–2018). Geneva: World Health Organization; 2018.
Snounou G, Viriyakosol S, Zhu Xin Ping, Jarra W, Pinheiro L, V E do Rosario, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–20.
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–7.
Ahmed A, Bararia D, Vinayak S, Yameen M, Biswas S, Dev V, et al. Plasmodium falciparum isolates in India exhibit a progressive increase in mutations associated with sulfadoxine-pyrimethamine resistance. Antimicrob Agents Chemother. 2004;48:879–89.
Laufer MK, Takala-Harrison S, Dzinjalamala FK, Stine OC, Taylor TE, Plowe CV. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J Infect Dis. 2010;202:801–8.
Frosch AEP, Laufer MK, Mathanga DP, Takala-Harrison S, Skarbinski J, Claassen CW, et al. Return of widespread chloroquine-sensitive Plasmodium falciparum to Malawi. J Infect Dis. 2014;210:1110–4.
Galatas B, Nhamussua L, Candrinho B, Mabote L, Cisteró P, Gupta H, et al. In vivo efficacy of chloroquine to clear asymptomatic infections in Mozambican adults: a randomized, placebo-controlled trial with implications for elimination strategies. Sci Rep. 2017;7:1356.
Balikagala B, Sakurai-Yatsushiro M, Tachibana SI, Ikeda M, Yamauchi M, Katuro OT, et al. Recovery and stable persistence of chloroquine sensitivity in Plasmodium falciparum parasites after its discontinued use in Northern Uganda. Malar J. 2020;19:76.
Madkhali AM, Abdulhaq AA, Atroosh WM, Ghzwani AH, Zain KA, Ghailan KY, et al. The return of chloroquine-sensitive Plasmodium falciparum parasites in Jazan region, southwestern Saudi Arabia over a decade after the adoption of artemisinin-based combination therapy: analysis of genetic mutations in the pfcrt gene. Parasitol Res. 2021;120:3771–81.
Ogouyèmi-Hounto A, Ndam NT, KindeGazard D, D’Almeida S, Koussihoude L, Ollo E, et al. Prevalence of the molecular marker of Plasmodium falciparum resistance to chloroquine and sulphadoxine/pyrimethamine in Benin seven years after the change of malaria treatment policy. Malar J. 2013;12:1147.
Patgiri SJ, Sarma K, Sarmah N, Bhattacharyya N, Sarma DK, Nimolia T, et al. Characterization of drug resistance and genetic diversity of Plasmodium falciparum parasites from Tripura. Northeast India Sci Rep. 2019;9:13704.
Ozarkar A, Kanyal A, Dass S, Deshpande P, Deobagkar D, Karmodiya K. Analysis of drug resistance marker genes of Plasmodium falciparum after implementation of artemisinin-based combination therapy in Pune district. India J Biosci. 2021;46:77.
Pathak A, Mårtensson A, Gawariker S, Sharma A, Diwan V, Purohit M, et al. Stable high frequencies of sulfadoxine-pyrimethamine resistance associated mutations and absence of K13 mutations in Plasmodium falciparum 3 and 4 years after the introduction of artesunate plus sulfadoxine-pyrimethamine in Ujjain, Madhya Pradesh. India Malar J. 2020;19:290.
Zomuanpuii R, Hmar CL, Lallawmzuala K, Hlimpuia L, Balabaskaran Nina P, Senthil KN. Epidemiology of malaria and chloroquine resistance in Mizoram, northeastern India, a malaria-endemic region bordering Myanmar. Malar J. 2020;19:95.
Awasthi G, Satya GBK, Das A. Pfcrt haplotypes and the evolutionary history of chloroquine-resistant Plasmodium falciparum. Mem Inst Oswaldo Cruz. 2012;107:129–34.
Das A. The distinctive features of Indian malaria parasites. Trends Parasitol. 2015;31:83–6.
Ranjit MR, Sahu U, Khatua CR, Mohapatra BN, Acharya AS, Kar SK. Chloroquine-resistant P. falciparum parasites and severe malaria in Orissa. Curr Sci. 2009;96:1608–11.
Fairhurst RM. Understanding artemisinin-resistant malaria: what a difference a year makes. Curr Opin Infect Dis. 2015;28:417–25.
Das S, Saha B, Hati AK, Roy S. Evidence of artemisinin-resistant Plasmodium falciparum malaria in Eastern India. N Engl J Med. 2018;379:1962–4.
Arya A, Kojom Foko LP, Chaudhry S, Sharma A, Singh V. Artemisinin-based combination therapy (ACT) and drug resistance molecular markers: a systematic review of clinical studies from two malaria endemic regions—India and sub-Saharan Africa. Int J Parasitol Drugs Drug Resist. 2021;15:43–56.
Chhibber-Goel J, Sharma A. Profiles of Kelch mutations in Plasmodium falciparum across South Asia and their implications for tracking drug resistance. Int J Parasitol Drugs Drug Resist. 2019;11:49–58.
Hemingway J, Shretta R, Wells TNC, Bell D, Djimdé AA, Achee N, et al. Tools and strategies for malaria control and elimination: what do we need to achieve a grand convergence in malaria? PLoS Biol. 2016;14:e1002380.
Mulenga MC, Sitali L, Ciubotariu II, Hawela MB, Hamainza B, Chipeta J, et al. Decreased prevalence of the Plasmodium falciparum Pfcrt K76T and Pfmdr1 and N86Y mutations post-chloroquine treatment withdrawal in Katete District. Eastern Zambia Malar J. 2021;20:329.
Kim J, Tan YZ, Wicht KJ, Erramilli SK, Dhingra SK, Okombo J, et al. Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature. 2019;576:315–20.
Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 2012;28:504–14.
Xu C, Wei Q, Yin K, Sun H, Li J, Xiao T, et al. Surveillance of antimalarial resistance Pfcrt, Pfmdr1 and Pfkelch13 polymorphisms in African Plasmodium falciparum imported to Shandong Province, China. Sci Rep. 2018. https://doi.org/10.1038/s41598-018-31207-w.
Oguike MC, Falade CO, Shu E, Enato IG, Watila I, Baba ES, et al. Molecular determinants of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum in Nigeria and the regional emergence of dhps 431V. Int J Parasitol Drugs Drug Resist. 2016;6:220–9.
McCollum AM, Poe AC, Hamel M, Huber C, Zhou Z, Shi YP, et al. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J Infect Dis. 2006;194:189–97.
Hyde JE. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 2002;4:165–74.
Ndiaye D, Daily JP, Sarr O, Ndir O, Gaye O, Mboup S, et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes in Senegal. Trop Med Int Health. 2005;10:1176–9.
Roper C, Pearce R, Bredenkamp B, Gumede J, Drakeley C, Mosha F, et al. Antifolate antimalarial resistance in Southeast Africa: a population-based analysis. Lancet. 2003;361:1174–81.
Wang P, Lee CS, Bayoumi R, Djimde A, Doumbo O, Swedberg G, et al. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroatesynthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol Biochem Parasitol. 1997;89:161–77.
Kun JFJ, Lehman LG, Lell B, Schmidt-Ott R, Kremsner PG. Low-dose treatment with sulfadoxine-pyrimethamine combinations selects for drug-resistant Plasmodium falciparum strains. Antimicrob Agents Chemother. 1999;43:2205–8.
Dunyo S, Ord R, Hallett R, Jawara M, Walraven G, Mesa E, et al. Randomised trial of chloroquine/sulphadoxine-pyrimethamine in Gambian children with malaria: impact against multidrug-resistant P. falciparum. PLoS Clin Trials. 2006. https://doi.org/10.1371/journal.pctr.0010014.
Picot S, Olliaro P, De Monbrison F, Bienvenu AL, Price RN, Ringwald P. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J. 2009;8:89.
Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–8.
Acknowledgements
RKR and MR thank the Director of ICMR-Regional Medical Research Centre Bhubaneswar, Odisha, India, for providing essential laboratory facilities during work. RKR is thankful to the Indian Council of Medical Research for the award of a Senior Research Fellowship (SRF). The authors thank all the laboratory technicians who worked hard during the collection of the P. falciparum field isolate from all over the state, particularly from the tribal inaccessible areas, and also great thanks to all the individuals who participated in this study.
Funding
The Indian Council of Medical Research-Senior Research Fellowship (ICMR-SRF) (Letter Number-Fellowship/25/2018-ECD-II, Dated-29/07/2018) to Ramakanta Rana (RKR) provided financial support for the research and preparation of the article, and study design, the collection, analysis, and interpretation of data.
Author information
Authors and Affiliations
Contributions
RKR, MR, and MSB conceptualized the study, and RKR, MSB, and MR collected the patient samples and obtained relevant malaria epidemiological details. RKR and SS did all laboratory work, and malaria parasite genotyping and PCR-sequencing experiments of the genes. RKR, AD, and NK carried out all the sequencing analysis and bioinformatics work and wrote the interpretations, RKR, AD, and MR wrote the manuscript with contributions from SS, NK, MSB, and SP. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The Institutional Human Ethics Committee of the ICMR-RMRC Bhubaneswar has approved the study. Before the enrolment, the purpose of the study was explained to the participants in the local language, and verbal consent was obtained for blood sample collection and testing. One consent from the adult patient (above18 year age) and consent for children less than 18-year age from their parents or head of the house hold members in case of no parents, as per ICMR guidelines; written informed consent was obtained from patients or the parents/guardians of children prior to blood collection.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Rana, R., Khan, N., Sandeepta, S. et al. Molecular surveillance of anti-malarial drug resistance genes in Plasmodium falciparum isolates in Odisha, India. Malar J 21, 394 (2022). https://doi.org/10.1186/s12936-022-04403-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-022-04403-3