Abstract
Background
Anti-malarial drug resistance is a severe challenge for eventual control and global elimination of malaria. Resistance to sulfadoxine-pyrimethamine (SP) increases as mutations accumulate in the Pfdhfr and Pfdhps genes. This study aimed to assess the polymorphisms and prevalence of mutation in these genes in the Plasmodium falciparum infecting migrant workers returning to Wuhan, China.
Methods
Blood samples were collected for 9 years (2011–2019). Parasite genomic DNA was extracted from blood spots on filter paper. The mutations were evaluated by nested PCR and sequencing. The single-nucleotide polymorphisms (SNPs) and haplotypes of the Pfdhfr and Pfdhps genes were analysed.
Results
Pfdhfr codon 108 showed a 94.7% mutation rate, while for Pfdhps, the rate for codon 437 was 79.0%. In total, five unique haplotypes at the Pfdhfr locus and 11 haplotypes at the Pfdhps locus were found while the Pfdhfr-Pfdhps combined loci revealed 28 unique haplotypes. A triple mutant (IRNI) of Pfdhfr was the most prevalent haplotype (84.4%). For Pfdhps, a single mutant (SGKAA) and a double mutant (SGEAA) were detected at frequencies of 37.8 and 22.3%, respectively. Among the combined haplotypes, a quadruple mutant (IRNI-SGKAA) was the most common, with a 30.0% frequency, followed by a quintuplet mutant (IRNI-SGEAA) with a frequency of 20.4%.
Conclusion
The high prevalence and saturation of Pfdhfr haplotypes and the medium prevalence of Pfdhps haplotypes demonstrated in the present data will provide support for predicting the status and progression of antifolate resistance in malaria-endemic regions and imported malaria in nonendemic areas. Additional interventions to evaluate and prevent SP resistance should be continuously considered.
Similar content being viewed by others
Background
Malaria is caused by the Plasmodium parasite, which is transmitted to human beings via the bites of infected female Anopheles mosquitoes. It is prevalent in the tropics and subtropics, particularly sub-Saharan Africa, as well as in Southeast Asia (SEA) and South America. In 2018, there were an estimated 228 million new cases of malaria, which was responsible for approximately 405,000 deaths [1]. Among them, pregnant women and children under 5 years old in Africa are thought to be the primary victims.
In the 1980s, sulfadoxine-pyrimethamine (SP) replaced chloroquine (CQ) as the front-line anti-malaria treatment when large-scale CQ resistance developed in sub-Saharan African countries. However, SP soon had to be replaced by artemisinin-based combination therapy (ACT) due to drug resistance. However, SP is still used for intermittent preventive treatment in infants (IPTi) and pregnant women (IPTp) during malaria-endemic regions, following the guidance of the World Health Organization (WHO) [2]. Furthermore, the administration of SP plus amodiaquine is applied for seasonal malaria chemoprevention (SMC) [3]. Currently, the emergence, development, and continuous dissemination of Plasmodium falciparum resistance to the anti-malarial drug is considered a significant global threat for malaria control and elimination strategies [4]. The development of drug resistance could be influenced by multiple factors, including mutation frequency, treatment costs, drug selection pressure, patient compliance, and host immunity [5, 6]. It is necessary to conduct molecular epidemiological surveillance and monitoring of drug-resistant P. falciparum parasites from disease-endemic to nonendemic areas. Molecular markers are a useful tool for confirming that parasites are drug-resistant.
For Plasmodium spp., enzymes involved in folate metabolism are interfered with by the antifolate anti-malarial drugs. Pyrimethamine acts as an inhibitor in P. falciparum dihydrofolate reductase (Pfdhfr) and sulfadoxine, targets the P. falciparum enzyme dihydropteroate synthase (Pfdhps) [7]. In vitro and in vivo studies have demonstrated resistance to SP is mainly mediated by mutations at codons Pfdhfr N51I, C59R, S108N, and I164L, and Pfdhps S436A, A437G, K540E, A581G, and A613S [8, 9]. SP resistance, with is very common, is accompanied by the accumulation of these mutations [10,11,12]. In particular, combinations of multiple mutations in both genes, such as the quadruple mutant carrying four partially resistant mutation in combination, are mainly comprised of the Pfdhfr triple mutant (N51I/C59R/S108N) and Pfdhps (A437G). The quintuple mutant genotype includes the Pfdhfr (N51I/C59R/S108N) and Pfdhps (A437G/K540E). The sextuple mutant consists of a triple mutant (N51I/C59R/S108N) in Pfdhfr and a triple mutant (A437G/ K540E/A581G) in Pfdhps, a combination that was called super resistant [13,14,15]. Multiple combinations of mutations can affect IPTi and IPTp treatment outcomes. Therefore, it is necessary not only to monitor the increase of mutations at a single site, but also to prevent the potential combination of more other multiple mutations.
This study investigated the prevalence of the mutant and wild-type alleles isolated from P. falciparum infecting migrant workers who have returned to Wuhan, central China, who all came from perennial transmission regions from 2011–2019. Such molecular surveillance will provide health authorities with valuable information for adopting efficient anti-malarial drugs in malaria-endemic regions in Africa and malaria nonendemic areas with imported malaria in China particularly Wuhan.
Methods
Samples collection
Blood samples were collected from P. falciparum-infected migrant patients with uncomplicated malaria in Wuhan of Hubei Province from 2011 to 2019. These samples were examined by microscopy with stained thick and thin blood smears and detected by rapid diagnostic tests (RDTs) for Pf-HRP2 and pLDH, as previously described [16,17,18]. RDTs, were carried out according to the manufacturer's manual (Wondfo, Guangzhou, China). Subsequently, two or three drops of blood were spotted on Whatman 3MM filter paper, air dried, and stored in an individually coded sealed plastic bag containing silica desiccant beads. The bags were stored at 4 °C until use. The study was approved by the Ethical Review Committee of the Hubei University of Medicine and Wuhan City Center for Disease Prevention and Control Ethics Committee. Informed consent was obtained and signed by all participants or their guardians before inclusion in the study and before sample collection.
Molecular procedures
Genomic DNA (gDNA) from dried blood spot samples was isolated using the TIANamp Blood DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) following the manufacturer’s recommendations. DNA samples were stored at − 20 °C for further genotyping.
To identify polymorphisms in the Pfdhfr (Gene ID: PF3D7_0417200) and Pfdhps (Gene ID: PF3D7_0810880) genes, purified gDNA templates were amplified by using nested PCR in the Mini MJ Thermal Cycler (Bio-Rad), following protocols described previously with minor changes [19, 20]. Briefly, primary PCR, was performed in a total of 20 μl containing 10 μl of 2 × Phusion PCR Master Mix (40 units/ml Phusion DNA polymerase, 400 μM deoxynucleoside triphosphate [dNTP] mixture, 2 × Phusion high-fidelity [HF] buffer, and 3 mM Mg2+), 1 μl of each primer (10 μM), and 2 μl of gDNA. For the second round, 1.0 μl of primary PCR products were amplified with a 50 μl reaction system. All PCR conditions conducted were as follows: predenaturation at 95 °C for 3 min, followed by 30 cycles of denaturing for 30 s at 95 °C, annealing for 30 s at 55 °C and extension at 72 °C for 30 s, plus a final extension at 72 °C for 5 min. The PCR product was electrophoresed on a 1.0% agarose gel stained with GelRed®Safe DNA Gel Stain (Boitium, USA) and visualized with a UV transilluminator. Sequencing was outsourced to Genewiz, Soochow, China, whereby for the Pfdhfr gene, the nested PCR products were purified and sequencing in reverse directions, while for the Pfdhps gene, the nested PCR products were purified and sequenced by Bidirectional DNA sequencing. Polymorphisms were analysed by creating consensus nucleotide sequences with the reference sequences in PlasmoDB using DNAStar (DNASTAR Inc., Madison, WI, USA).
Statistical analysis
The frequencies of single-nucleotide polymorphisms (SNPs) and haplotypes were calculated as the percentage of the number of successful sequencing samples. Mixed infections containing wild-type and mutation were excluded from further analysis. The comparison and trend estimation of haplotypes were assessed by unpaired T-test and linear regression analysis respectively with GraphPad Prism 5.
Results
Molecular surveillance of Pfdhfr and Pfdhps mutations
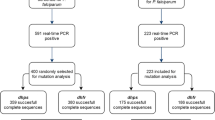
In total, 303 isolates from 2011 to 2019 were included for the analysis of Pfdhfr and Pfdhps mutations. Table 1 summarizes the different SNPs observed in the samples collected over these years. For Pfdhfr, mutations at different loci, including N51I, C59R, S108N, and I164L, were investigated. Of the 300 samples successfully genotyped for the Pfdhfr gene, 90.7% (272/300) harboured the mutant allele N51I, while six samples (2.0%) had a mixed type. For the Pfdhfr codon at 59, 84.7% (254/300) samples had the mutant allele C59R, and 9 (3%) had mixed genotypes. At codon 108, 94.7% (284/300) harboured a mutation, 3.0% (9/300) were wild type, and 2.3% (7/300) mixed infection were found. Only 1 sample out of 300 (0.3%) derived from Myanmar in 2011 had the highly drug resistant mutation I164L. However, we did not observe any mutation at codon 50 of Pfdhfr. For Pfdhps, mutations at different loci, including S436A, A437G, K540E, A581G, and A613S, were surveyed. Of the 290 (290/303, 95.7%) successful genotyped samples, 27.2% of the isolates harboured S436A, 79.0% harboured A437G, 21.7% harboured K540E, 9.0% harboured A581G and 11.0% harboured A613S. For wild type, 66.0% of the isolates harboured S436A, 13.1% A437G, 70.3% K540E, 85.5% A581G and 89.0% A613S, respectively. For mixed genotypes, 6.9% of the isolates harboured S436A, 7.9% harboured A437G, 7.9% harboured K540E, 5.5% harboured A581G and 0.3% harboured A613S.
Prevalence of the Pfdhfr and Pfdhps haplotypes
In total, five unique haplotypes at the Pfdhfr locus and 11 haplotypes at the Pfdhps locus were found. The prevalence of the Pfdhfr and Pfdhps haplotypes in different years and various geographical regions are illustrated in Fig. 1. The Pfdhfr mutations were the most prevalent, with the triple mutation (IRNI) in almost 84.4% (238/282). Compared to the haplotypes of NCSI, ICNI, and NRNI, the IRNI displayed a higher prevalence during 2011–2019 (P < 0.0001, P < 0.0001, and P < 0.0001, respectively) (Fig. 1a). As the predominant haplotype, the IRNI was found in all geographical areas in Africa and SEA (Fig. 1b). For NCSI (wild type), the frequency has increased from none in 2011 and 2012 to 9.1% (3/33) in 2018, and then reduced to 2.5% (1/40) in 2019. The relative frequencies of the ICNI mutations decreased from 16.67% (1/6) in 2011 to 0 in 2012 and increased to 14.3% (6/42) in 2013; finally stabilized at 15.0% (6/40) in 2019. For NRNI, there was only a marginal difference over these years (F = 0.6316, P = 0.4529). The allele with quadruple mutations (IRNL), which conferred a high level of resistance to antifolates, was only found at a low frequency (0.3%, 1/300) in 2011. For Pfdhps, the predominant haplotype was single-mutant SGKAA (37.8%, 90/238). It was increased from 14.3% (1/7) in 2011 to 30.0% (9/30) in 2019 (Fig. 1c). Although a decreasing trend was observed in the prevalence of the quadruple-mutant AGKGS from 18.5% in 2012 to 9.8% in 2016 and finally to 0% in 2019, these differences were not statistically significant (F = 0.0063, P = 0.9391). Similarly, the prevalence of the Pfdhps wild type SAKAA genotype has decreased from 42.9% in 2011 to 3.33% in 2019, but it was not statistically significant (F = 4.456, P = 0.0727). The Pfdhps double-mutant AGKAA genotype decreased from 14.3% in 2011 to 7.3% in 2016 and finally increased to 16.7% in 2019 (F = 0.6879, P = 0.4342). A concomitant increase in the prevalence of the Pfdhps double-mutant SGEAA genotype was observed, increasing in prevalence from 3.7% in 2012 to 33.3% in 2019, which was statistically significant (F = 9.034, P = 0.0198) (Fig. 1c). As the predominant haplotype, the SGKAA was mostly found in WA, CA, and SA with a prevalence of 93.4, 47.5, and 42.9%, respectively (Fig. 1d). SGEAA, was mostly distributed in SA (36.7%), EA (76.2%), and CA (17.0%) (Fig. 1d).
Observed haplotypes of Pfdhfr and Pfdhps in different years and areas. a The haplotypes of Pfdhfr in different years. b The haplotypes of Pfdhfr in different regions. c The haplotypes of Pfdhps in different years. d The haplotypes of Pfdhps in different areas. e The combined haplotypes of Pfdhfr-Pfdhps in different years. f The combined haplotypes of Pfdhfr-Pfdhps in different regions. The WA, SA, CA, EA, NA, and SEA represent West Africa, South Africa, Central Africa, East Africa, North Africa, and Southeast Asia, respectively
Prevalence of Pfdhfr and Pfdhps combination haplotypes
A total of 230 samples were subjected to combined haplotypes analysis. The 28 haplotypes were verified by combining both genes. The most common haplotype was IRNI-SGKAA with a 30.0% (69/230) frequency, followed by IRNI-SGEAA (20.4%, 47/230), IRNI-AGKAA (9.8%, 22/230), and IRNI-AGKGS (7.8%, 18/230). Moreover, the frequency distribution of the different Pfdhfr-Pfdhps haplotypes was compared among the analysed samples collected at different times. The results showed that the most prevalent haplotype observed during the study period was IRNI-SGKAA, which also remained the most frequent one in WA (32.7%), SA (23.4%), and CA (44.4%) (Fig. 1e). For IRNI-SGEAA, an increasing trend was detected during 2011–2016, and 2017–2019, respectively (Fig. 1e). For regional distribution, IRNI-SGKAA was mainly distributed in WA (32.7%), CA (44.4%), and SA (23.4%) (Fig. 1F). Similarly, IRNI-SGEAA was mainly found in EA (71.4%) and SA (31.9%), followed by CA (16.7%) and WA (7.1%) (Fig. 1f). An additional 20 minor haplotypes with a prevalence of less than 2% constituted only 9.1% (21/227) of the overall haplotypes (Additional file 1: Table S1 and Additional file 2: Table S2).
Discussion
Globally, strategies for malaria control have substantially reduced the disease burden in the last few decades. Soon afterward, several nations in Asia (particularly China), Africa, and Latin America began advancing towards malaria elimination [21,22,23]. However, imported malaria from Africa and SEA has affected and delayed the progress of malaria elimination in China. Furthermore, drug-resistant P. falciparum parasites will become a significant challeng influencing the process of malaria control, elimination, and eradication. The SNPs in the Pfdhfr and Pfdhps genes are linked to the failure of SP treatment against uncomplicated P. falciparum malaria and have been documented in Africa and SEA for several decades [10, 11]. However, there are no such data to support drug policies in nonendemic areas with imported malaria in China particularly Wuhan [24]. To determine whether parasites carrying these polymorphisms exist in Wuhan, molecular surveillance were conducted targeting Pfdhfr and Pfdhps gene polymorphisms in imported clinical isolates.
For Pfdhfr, the critical event in the development of pyrimethamine resistance is a mutation in codon 108 that changes serine (S) to asparagine (N), resulting in partial pyrimethamine resistance. Further mutations at N51I and/or C59R increase the level of pyrimethamine resistance [25]. Under continuous pyrimethamine selective drug pressure, the SNP adaptations in our data have also followed this rule. The current survey demonstrated an extremely high prevalence (> 84%) of three mutations (N51I, C59R, and S108N) in P. falciparum clinical isolates imported from Africa and SEA. In Africa, Pfdhfr nonsynonymous polymorphisms have also been reported at high frequencies in isolates from Uganda [26], Angola [27], the Democratic Republic of the Congo [12], Nigeria [28], and Sierra Leone [29]. Parasitic infections carrying the Pfdhfr triple mutant (IRNI) are significantly more likely to be resistant to SP treatment than infections with fewer Pfdhfr mutations [30]. Most of the isolates in our dataset had the triple mutant allele IRNI (84.4%, 238/282), indicating that pyrimethamine resistance remains at a relatively high level in Africa. However, no mutations in codons 50 and 164 of Pfdhfr were detected in samples collected on the African continent. An additional mutation, I164L, confers an elevated level of pyrimethamine resistance that could render SP invalid [27]. Although the Pfdhfr I164L mutation was first reported from Kenya [31] and then was found in Madagascar [32] and the Central African Republic [33], it was not detected in Africa in the present study. Furthermore, only one sample (0.3%, 1/300) with the I164L mutation was found in 2011 from SEA (Myanmar).
For Pfdhps, as the key mutation associated with sulfadoxine resistance, a single amino acid residue changes from alanine (A) to glycine (G) at codon 437 of Pfdhps [34]. The A437G selection by SP has been previously described during IPTi [35]. As the most frequent mutation of Pfdhps, our findings illustrate the high prevalence of A437G at 79.0% (229/290), which has also been reported to be nearly at a saturation level in most African countries [27]. Furthermore, a higher proportion of A437G has been detected at 75.6% in Gabon [36], 87.9% in Kenya [37], 97.6% (1416/1451) in Congo [12], and 96.4% (27/28) in Nigeria [28]. Thus, it needs to be kept in mind that a high prevalence of SP-resistant parasites is present in these regions. More attention should be given to SP drug resistance surveillance, both in these countries and in nonedemic areas, particularly Wuhan, which is influenced by imported malaria from endemic areas. Compared to the high prevalence of Pfdhfr mutations, a low prevalence (< 30%) of four mutant alleles (S436A, K540E, A581G, and A613S) in Pfdhps was detected. It has been reported that Pfdhps K540E has a low prevalence in Central and West Africa [9, 14, 28, 33]. In contrast, the K540E mutation is common in East Africa [38, 39], similar to our findings. The Pfdhps A581G and A613S/T mutations have been detected at a low prevalence in WA and EA, but a rapid emergence of these mutations has been described in Kenya and Uganda [25, 33, 40]. Apart from Nigeria and Cameroon, these mutations have not been found in CA [41]. In Cameroon, there is an increasing trend in the prevalence of the Pfdhps A581G and A613S mutations [33]. In the current study, the prevalence of A581G and A613S were generally consistent with these observations in Africa.
Parasites carrying all five mutations, the Pfdhfr triple mutant (N51I + C59R + S108N) and the Pfdhps double mutant (A437G and K540E), commonly called the quintuple mutation (IRNI-SGEAA), have been strongly associated with SP treatment failure in sub-Saharan Africa [42,43,44,45]. Alarmingly, the present study found 18.54% of tested isolates harboured fully resistant (IRNI-SGEAA), which is common in EA [34, 46]. Additional mutations in Pfdhfr I164L and Pfdhps A581G have been associated with a high level of SP resistance and failure [47]. However, a quintuple mutant named “super-resistant genotypes” is linked with a more than triple enhancement of therapeutic failure [47]. The IRNI-SGEGA forming the sextuple haplotype has been connected with an optimal resistance effect, referred to as the “super-resistant genotype” [37]. Three isolates of such genotype are found in our data, which is of concern. In addition, it is noteworthy that the other combined haplotypes, including the quintuple mutant (IRNI-AGKAA, IRNI-SGKAS, ICNI-SGEGA), the sextuple mutant (IRNI-AGKAS, NRNI-AGKGS, IRNI-AGEAA), and the septuple mutant (IRNI-AGKGS, IRNL-SGEGA) were also detected in the current data. Previous studies revealed that IRNI-SGEGA, IRNI-AGEAA has been highly associated with a lack of IPTp-SP efficacy [48]. It was illustrated that such genotypes were widely distributed in Tanzania, in line with our study, where one isolate harboured the sextuple haplotype (IRNI-SGEGA), which happened to come from Tanzania [48]. Interestingly, the septuple mutant haplotype (IRNI-AGKGS) accounts for a certain proportion of our findings, which is similar to a previous study reported in Nigeria [3]. This demonstrates that SP resistance remains at a moderate level in Africa. Although SP is recommended as an effective anti-malarial drug used for the vulnerable population [35], more attention needs to be paid to these mutations profiles.
Conclusions
In conclusion, the present study reports the persistence of P. falciparum parasites with Pfdhfr and Pfdhps mutations associated with SP resistance in migrant workers returning from Africa and SEA to Wuhan, central China. These findings provide fundamental prevalence data that enable a policy-making organization to directly determine the best measures and strategies for malaria control and elimination.
Availability of data and materials
The datasets analysed in this study are available from the corresponding author on reasonable request.
Abbreviations
- A:
-
Alanine
- G:
-
Glycine
- N:
-
Asparagine
- S:
-
Serine
- ACTs:
-
Artemisinin-based combination therapies
- CQ:
-
Chloroquine
- gDNA:
-
Genomic DNA
- Pfdhps :
-
P. falciparum Dihydropteroate synthase
- Pfdhfr :
-
P. falciparum Dihydrofolate reductase
- IPTi:
-
Intermittent preventive treatment in infants
- IPTp:
-
Intermittent preventive treatment pregnant women
- RDTs:
-
Rapid diagnostic tests
- Pf-HRP2:
-
P. falciparum Specific Histidine-rich protein 2
- pLDH:
-
Plasmodium lactate dehydrogenase
- SMC:
-
Seasonal Malaria Chemoprevention
- SEA:
-
Southeast Asia
- SP:
-
Sulfadoxine-pyrimethamine
- SNPs:
-
Single-nucleotide polymorphisms
References
World Health Organization. World malaria report 2019. Geneva: World Health Organization; 2019.
World Health Organization, Global Malaria Programme. WHO policy brief for the implementation of intermittent preventive treatment of malaria in pregnancy using sulfadoxine-pyrimethamine (IPTp-SP). Geneva: World Health Organization; 2013.
Oguike MC, Falade CO, Shu E, Enato IG, Watila I, Baba ES, et al. Molecular determinants of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum in Nigeria and the regional emergence of dhps 431V. Int J Parasitol Drugs Drug Resist. 2016;6:220–9.
Artimovich E, Schneider K, Taylor TE, Kublin JG, Dzinjalamala FK, Escalante AA, et al. Persistence of sulfadoxine-pyrimethamine resistance despite reduction of drug pressure in Malawi. J Infect Dis. 2015;212:694–701.
Petersen I, Eastman R, Lanzer M. Drug-resistant malaria: molecular mechanisms and implications for public health. FEBS Lett. 2011;585:1551–62.
Gupta H, Macete E, Bulo H, Salvador C, Warsame M, Carvalho E, et al. Drug-resistant polymorphisms and copy numbers in Plasmodium falciparum, Mozambique, 2015. Emerg Infect Dis. 2018;24:40–8.
Corredor V, Murillo C, Echeverry DF, Benavides J, Pearce RJ, Roper C, et al. Origin and dissemination across the Colombian Andes mountain range of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54:3121–5.
Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RA, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–8.
Ruh E, Bateko JP, Imir T, Taylan-Ozkan A. Molecular identification of sulfadoxine-pyrimethamine resistance in malaria infected women who received intermittent preventive treatment in the Democratic Republic of Congo. Malar J. 2018;17:17.
Mita T, Tanabe K, Takahashi N, Culleton R, Ndounga M, Dzodzomenyo M, et al. Indigenous evolution of Plasmodium falciparum pyrimethamine resistance multiple times in Africa. J Antimicrob Chemother. 2009;63:252–5.
Pathak A, Martensson A, Gawariker S, Mandliya J, Sharma A, Diwan V, et al. Characterization of drug resistance associated genetic polymorphisms among Plasmodium falciparum field isolates in Ujjain, Madhya Pradesh. India Malar J. 2014;13:182.
Nkoli Mandoko P, Rouvier F, Matendo Kakina L, Moke Mbongi D, Latour C, Losimba Likwela J, et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine/pyrimethamine in the Democratic Republic of the Congo: emergence of highly resistant pfdhfr/pfdhps alleles. J Antimicrob Chemother. 2018;73:2704–15.
Moussiliou A, De Tove YS, Doritchamou J, Luty AJ, Massougbodji A, Alifrangis M, et al. High rates of parasite recrudescence following intermittent preventive treatment with sulphadoxine-pyrimethamine during pregnancy in Benin. Malar J. 2013;12:195.
Berzosa P, Esteban-Cantos A, Garcia L, Gonzalez V, Navarro M, Fernandez T, et al. Profile of molecular mutations in pfdhfr, pfdhps, pfmdr1, and pfcrt genes of Plasmodium falciparum related to resistance to different anti-malarial drugs in the Bata District (Equatorial Guinea). Malar J. 2017;16:28.
Naidoo I, Roper C. Mapping “partially resistant”, “fully resistant”, and “super resistant” malaria. Trends Parasitol. 2013;29:505–15.
Maltha J, Gamboa D, Bendezu J, Sanchez L, Cnops L, Gillet P, et al. Rapid diagnostic tests for malaria diagnosis in the Peruvian Amazon: impact of pfhrp2 gene deletions and cross-reactions. PLoS ONE. 2012;7:e43094.
Yao Y, Wu K, Xu M, Yang Y, Zhang Y, Yang W, et al. Surveillance of genetic variations associated with antimalarial resistance of Plasmodium falciparum Isolates from returned migrant workers in Wuhan, Central China. Antimicrob Agents Chemother. 2018;62:e02387-e2417.
Mehlotra RK, Howes RE, Cramer EY, Tedrow RE, Rakotomanga TA, Ramboarina S, et al. Plasmodium falciparum parasitemia and band sensitivity of the SD Bioline Malaria Ag P.f/Pan rapid diagnostic test in Madagascar. Am J Trop Med Hyg. 2019;100:1196–201.
Pearce RJ, Drakeley C, Chandramohan D, Mosha F, Roper C. Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of northern Tanzania. Antimicrob Agents Chemother. 2003;47:1347–54.
Jiang T, Chen J, Fu H, Wu K, Yao Y, Eyi JUM, et al. High prevalence of Pfdhfr-Pfdhps quadruple mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates from Bioko Island. Equatorial Guinea Malar J. 2019;18:101.
O’Meara WP, Mangeni JN, Steketee R, Greenwood B. Changes in the burden of malaria in sub-Saharan Africa. Lancet Infect Dis. 2010;10:545–55.
Lai S, Li Z, Wardrop NA, Sun J, Head MG, Huang Z, et al. Malaria in China, 2011–2015: an observational study. Bull World Health Organ. 2017;95:564–73.
World Health Organization. World Malaria Report 2018. Geneva: World Health Organization; 2018. p. 210.
Xia J, Huang X, Sun L, Zhu H, Lin W, Dong X, Wu D, et al. Epidemiological characteristics of malaria from control to elimination in Hubei Province, China, 2005–2016. Malar J. 2018;17:81.
Spalding MD, Eyase FL, Akala HM, Bedno SA, Prigge ST, Coldren RL, et al. Increased prevalence of the pfdhfr/phdhps quintuple mutant and rapid emergence of pfdhps resistance mutations at codons 581 and 613 in Kisumu. Kenya Malar J. 2010;9:338.
Mbonye AK, Birungi J, Yanow SK, Shokoples S, Malamba S, Alifrangis M, et al. Prevalence of Plasmodium falciparum resistance markers to sulfadoxine-pyrimethamine among pregnant women receiving intermittent preventive treatment for malaria in Uganda. Antimicrob Agents Chemother. 2015;59:5475–82.
Kaingona-Daniel EP, Gomes LR, Gama BE, Almeida-de-Oliveira NK, Fortes F, Menard D, et al. Low-grade sulfadoxine-pyrimethamine resistance in Plasmodium falciparum parasites from Lubango. Angola Malar J. 2016;15:309.
Esu E, Tacoli C, Gai P, Berens-Riha N, Pritsch M, Loescher T, et al. Prevalence of the Pfdhfr and Pfdhps mutations among asymptomatic pregnant women in Southeast Nigeria. Parasitol Res. 2018;117:801–7.
Smith SJ, Kamara ARY, Sahr F, Samai M, Swaray AS, Menard D, et al. Efficacy of artemisinin-based combination therapies and prevalence of molecular markers associated with artemisinin, piperaquine and sulfadoxine-pyrimethamine resistance in Sierra Leone. Acta Trop. 2018;185:363–70.
Khattak AA, Venkatesan M, Jacob CG, Artimovich EM, Nadeem MF, Nighat F, et al. A comprehensive survey of polymorphisms conferring anti-malarial resistance in Plasmodium falciparum across Pakistan. Malar J. 2013;12:300.
McCollum AM, Poe AC, Hamel M, Huber C, Zhou Z, Shi YP, et al. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J Infect Dis. 2006;194:189–97.
Andriantsoanirina V, Bouchier C, Tichit M, Jahevitra M, Rabearimanana S, Randrianjafy R, et al. Origins of the recent emergence of Plasmodium falciparum pyrimethamine resistance alleles in Madagascar. Antimicrob Agents Chemother. 2010;54:2323–9.
Chauvin P, Menard S, Iriart X, Nsango SE, Tchioffo MT, Abate L, et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine/pyrimethamine in pregnant women in Yaounde, Cameroon: emergence of highly resistant pfdhfr/pfdhps alleles. J Antimicrob Chemother. 2015;70:2566–71.
Braun V, Rempis E, Schnack A, Decker S, Rubaihayo J, Tumwesigye NM, et al. Lack of effect of intermittent preventive treatment for malaria in pregnancy and intense drug resistance in western Uganda. Malar J. 2015;14:372.
Ruperez M, Gonzalez R, Mombo-Ngoma G, Kabanywanyi AM, Sevene E, Ouedraogo S, et al. Mortality, morbidity, and developmental outcomes in infants born to women who received either mefloquine or sulfadoxine-pyrimethamine as intermittent preventive treatment of malaria in pregnancy: a cohort study. PLoS Med. 2016;13:e1001964.
Bouyou-Akotet MK, Tshibola ML, Mawili-Mboumba DP, Nzong J, Bahamontes-Rosa N, Tsoumbou-Bakana G, et al. Frequencies of dhfr/dhps multiple mutations and Plasmodium falciparum submicroscopic gametocyte carriage in Gabonese pregnant women following IPTp-SP implementation. Acta Parasitol. 2015;60:218–25.
Lucchi NW, Okoth SA, Komino F, Onyona P, Goldman IF, Ljolje D, et al. Increasing prevalence of a novel triple-mutant dihydropteroate synthase genotype in Plasmodium falciparum in western Kenya. Antimicrob Agents Chemother. 2015;59:3995–4002.
Kateera F, Nsobya SL, Tukwasibwe S, Hakizimana E, Mutesa L, Mens PF, et al. Molecular surveillance of Plasmodium falciparum drug resistance markers reveals partial recovery of chloroquine susceptibility but sustained sulfadoxine-pyrimethamine resistance at two sites of different malaria transmission intensities in Rwanda. Acta Trop. 2016;164:329–36.
Tumwebaze P, Tukwasibwe S, Taylor A, Conrad M, Ruhamyankaka E, Asua V, et al. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis. 2017;215:631–5.
Naidoo I, Roper C. Drug resistance maps to guide intermittent preventive treatment of malaria in African infants. Parasitology. 2011;138:1469–79.
Sutherland CJ, Fifer H, Pearce RJ, Bin Reza F, Nicholas M, Haustein T, et al. Novel pfdhps haplotypes among imported cases of Plasmodium falciparum malaria in the United Kingdom. Antimicrob Agents Chemother. 2009;53:3405–10.
Greenhouse B, Slater M, Njama-Meya D, Nzarubara B, Maiteki-Sebuguzi C, Clark TD, et al. Decreasing efficacy of antimalarial combination therapy in Uganda is explained by decreasing host immunity rather than increasing drug resistance. J Infect Dis. 2009;199:758–65.
Andrews KG, Lynch M, Eckert E, Gutman J. Missed opportunities to deliver intermittent preventive treatment for malaria to pregnant women 2003–2013: a systematic analysis of 58 household surveys in sub-Saharan Africa. Malar J. 2015;14:521.
Shah M, Omosun Y, Lal A, Odero C, Gatei W, Otieno K, et al. Assessment of molecular markers for anti-malarial drug resistance after the introduction and scale-up of malaria control interventions in western Kenya. Malar J. 2015;14:75.
Ravenhall M, Benavente ED, Mipando M, Jensen AT, Sutherland CJ, Roper C, et al. the impact of sustained sulfadoxine/pyrimethamine use upon the Plasmodium falciparum population in Malawi. Malar J. 2016;15:575.
Matondo SI, Temba GS, Kavishe AA, Kauki JS, Kalinga A, van Zwetselaar M, et al. High levels of sulphadoxine-pyrimethamine resistance Pfdhfr-Pfdhps quintuple mutations: a cross sectional survey of six regions in Tanzania. Malar J. 2014;13:152.
Rouhani M, Zakeri S, Pirahmadi S, Raeisi A, Djadid ND. High prevalence of pfdhfr-pfdhps triple mutations associated with anti-malarial drugs resistance in Plasmodium falciparum isolates seven years after the adoption of sulfadoxine-pyrimethamine in combination with artesunate as first-line treatment in Iran. Infect Genet Evol. 2015;31:183–9.
Baraka V, Ishengoma DS, Fransis F, Minja DT, Madebe RA, Ngatunga D, et al. High-level Plasmodium falciparum sulfadoxine-pyrimethamine resistance with the concomitant occurrence of septuple haplotype in Tanzania. Malar J. 2015;14:439.
Acknowledgements
The authors would like to thank the Department of Schistosomiasis and Endemic Diseases, Wuhan Center for Disease Prevention and Control, and all participants who have contributed their blood samples.
Funding
This study was supported by the Foundation for Innovative Research Team of Hubei University of Medicine (Grant Number FDFR201603) and the National Natural Science Foundation of China (Grant Number 81802046).
Author information
Authors and Affiliations
Contributions
JL conceived and designed the experiments. KW coordinated the field collections of patient isolates. KW carried out a microscopic examination and RDTs. TTJ, WJC, and YY performed the experiments. JL, TTJ, WJC, and HBT analysed the data. JL and TTJ wrote the paper. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The current study was approved by the ethics committees of the Hubei University of Medicine and Wuhan City Center for Disease Prevention and Control Ethics Committee. Informed consent was obtained from all participated individuals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Haplotypes distribution of Pfdhfr and Pfdhps in a different country during 2011-2019.
Additional file 2: Table S2.
The combined haplotypes distribution of Pfdhfr and Pfdhps in different years and areas.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jiang, T., Cheng, W., Yao, Y. et al. Molecular surveillance of anti-malarial resistance Pfdhfr and Pfdhps polymorphisms in African and Southeast Asia Plasmodium falciparum imported parasites to Wuhan, China. Malar J 19, 434 (2020). https://doi.org/10.1186/s12936-020-03509-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-020-03509-w