Abstract
Background
Few studies have been conducted in Pakistan to determine the efficacy of chloroquine and sulphadoxine-pyrimethamine (SP), which remain in use as treatment for Plasmodium vivax and in combination with artesunate to treat Plasmodium falciparum, respectively. In this study, samples from several sites across Pakistan were characterized to determine prevalence of molecular resistance markers in the P. falciparum chloroquine resistance transporter (pfcrt), multidrug resistance (pfmdr1), dihydrofolate reductase (pfdhfr) and dihydropteroate synthase ( pfdhps) genes and the origin of chloroquine-resistant P. falciparum parasites.
Methods
Microscopy-confirmed malaria parasite-positive blood samples from 801 patients across the country were collected in 2011. Of these, 171 infections were identified by polymerase chain reaction (PCR) as P. falciparum and analysed by pyrosequencing for mutations conferring chloroquine resistance (pfcrt codons 72–76), multidrug resistance (pfmdr1 N86Y, Y184F, S1034C, N1042D and D1246Y), pyrimethamine resistance (pfdhfr, C50R, N51I, C59R, S108N and I164L) and sulphadoxine resistance (pfdhps, S436A, A437G, K540E, A581G and A613T/S). pfmdr1 gene copy number variation was determined by real-time PCR, and microsatellites flanking the pfcrt locus were typed to determine the origin of the chloroquine-resistant haplotype.
Results
The pfcrt K76T mutation was found in all samples as part of the S72/V73/M74/N75/T76 (SVMNT) haplotype. Microsatellites flanking pfcrt showed high similarity to the signature found in India and Papua New Guinea. pfmdr1 N86Y was found in 20% of samples and all samples harboured a single copy of the pfmdr 1 gene. The pfdhfr double mutation C59R + S108N was present in 87% of samples while the pfdhfr triple mutant (N51I + C59R + S108N) was not detected. Pfdhps A437G was found in 60% of samples. Pure pfdhps K540E was rare, at 4%, but mixed genotype 540 K/E was found in 77% of samples. Similarly, pure pfdhps A581G was found in 4% of the isolates while mixed 581A/G was found in 39% of samples.
Conclusions
These results suggest an emerging problem with multidrug resistant P. falciparum in Pakistan. The chloroquine resistance genotype has reached complete fixation in the population, with a microsatellite pattern indicative of a selective sweep. Moreover, the prevalence of mutations in both pfdhfr and pfdhps, albeit without the presence of the pfdhfr triple mutant, indicates that continued monitoring is warranted to assess whether SP remains efficacious as a partner drug for artesunate for the treatment of P. falciparum.
Similar content being viewed by others
Background
In Pakistan, an estimated 500,000 episodes of malaria infection occur annually [1]. Although the majority of these cases is attributed to Plasmodium vivax, studies indicate that Plasmodium falciparum in Pakistan has been on the rise over the past few decades [2]. The proportion of malaria infections attributed to P. falciparum rose from 34 to 54% between 1987 and 1990 in north-west Pakistan [3], and the frequency of P. falciparum among microscopy-positive cases rose from 45% in 1995 to 68% in 2006 in the provinces of Balochistan and Sindh [4, 5]. In 2010, out of a total of 240,591 reported malaria cases in Pakistan, 73,857 (31%) were P. falciparum[6].
The first-line treatment of uncomplicated malaria of undetermined species in Pakistan is chloroquine. Chloroquine-resistant P. falciparum was first reported in Pakistan in 1984 [3] and subsequent in vivo studies have confirmed that chloroquine is not an efficacious treatment for P. falciparum in Central Asia. A survey conducted in western Pakistan in Afghan settlements during 1994–1995 reported in vivo chloroquine resistance ranging from 18 to 62% [7]. Both standard and extended-dose courses of chloroquine resulted in high levels of treatment failure in clinical studies conducted in Afghani refugees in Pakistan [8]. Other clinical studies conducted in refugee communities throughout the country indicated that chloroquine resistance increased as much as 15% in exposed populations in a single year, rising five- to six-fold between 1982 and 1992 [2].
In response to the declining efficacy of chloroquine, the World Health Organization (WHO) recommended artesunate plus sulphadoxine-pyrimethamine (AS + SP) as the first-line choice of treatment for uncomplicated P. falciparum malaria in Pakistan [9]. However, a 56% treatment failure rate with SP monotherapy was reported in Balochistan province in a study conducted between 2001 and 2005 [10], indicating that SP may be compromised as an effective partner drug.
Molecular surveillance of drug resistance has been used extensively to monitor sensitivity to chloroquine and SP. A point mutation causing an amino acid change from K to T at codon 76 of the P. falciparum chloroquine resistance transporter gene (pfcrt) confers resistance to chloroquine in vitro[11, 12] and is strongly associated with treatment failure in vivo[13]. The role of these mutations in chloroquine resistance has also been confirmed by transfection studies [11]. A small number of independent pfcrt resistant lineages has been found: two in South America, one in Papua New Guinea [14] which has also been found in Asia [15, 16], one observed in the Philippines, and one in Southeast Asia which spread to Africa [14, 17].
Mutations in the dihydropteroate synthase (pfdhps) and dihydrofolate reductase (pfdhfr) genes of P. falciparum confer resistance to SP. Point mutations at positions N51I, C59R, S108N and I164L in pfdhfr are associated with pyrimethamine resistance in vitro[18–20] and treatment failure in vivo[21–23]. Resistance to sulfas is conferred by mutations at codons A437G, K540E, A581G and A613T/S of the pfdhps gene, confirmed by in vitro[24–28] and in vivo[22–24] studies. Resistance to SP occurs in a step-wise fashion, with an increasing number of mutations in pfdhfr and pfdhps contributing to an increased risk of treatment failure [29]. Infections carrying the pfdhfr triple mutant (S108N + N51I + C59R) are significantly more likely to fail SP treatment than infections with fewer pfdhfr mutations [30], and the pfdhfr/pfdhps quintuple mutant (pfdhfr triple mutant S108N + N51I + C59R plus the pfdhps double mutant A437G + K540E) is highly predictive of clinical failure in Africa [23, 29, 31].
Elevated copy number of the pfmdr 1 gene is associated with susceptibility of P. falciparum to artemisinin, mefloquine, and lumefantrine [32–34], and has been used as a surveillance tool for artesunate-mefloquine resistance in Southeast Asia [33, 35]. Mutations in the pfmdr1 gene leading to the substitution of amino acids including N86Y, Y184F, S1034C, N1042D, and D1246Y also modulate in vitro susceptibility to a number of drugs [36], although their relevance in conferring clinical resistance remains uncertain.
In Pakistan, previous molecular surveys of drug resistance have focused on one or two sites in a small number of geographic regions. In these surveys, the most recent of which was conducted in 2007, high levels of pfcrt 76T have been reported [1, 15, 37], and multiple mutations in the pfdhfr and pfdhps genes have been found [1, 37, 38]. In this study, the prevalence of molecular markers of anti-malarial resistance was investigated in a comprehensive survey over 14 sites in four provinces of Pakistan and in the capital city of Islamabad. Collected in 2011, these samples provide information on the current distribution of drug-resistant genotypes in the pfcrt, pfmdr1, pfdhfr and pfdhps genes five years after the implementation of artesunate + SP as the first-line treatment for uncomplicated P. falciparum.
Methods
Study sites and ethics
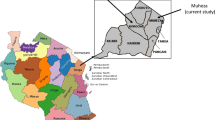
Government and private hospitals in 25 cities from four provinces with the highest burden of malaria representing all four provinces (Khyber Pakhtunkhwa province, Sindh province, Balochistan province and Punjab province) and a hospital in the capital city, Islamabad, were invited to participate in the molecular survey in 2011. Although the Federally Administered Tribal Areas have among the highest burdens of malaria in the country, they were excluded from this study because political instability and violence [39] make it difficult to establish sample collection. Of the facilities that were contacted, hospitals in 14 cities shared samples and were included in this study (Figure 1). Symptomatic patients of all age groups were enrolled and 3 mL of venous blood was drawn from each patient. Consent was obtained from patients or from children’s parents or guardians. Information on patient age and gender was also collected. The study was approved by the institutional review board of Quaid-i-Azam University, Islamabad, Pakistan.
Microscopy and sample collection
Thick and thin blood films of patients with suspected malaria were stained with 10% Giemsa solution and examined at 1,000 × magnification under oil immersion by a laboratory technician or technologist trained in malaria diagnosis according to WHO guidelines [40]. Approximately 50 μl of each blood sample from 801 malaria microscopy-positive and 30 microscopy-negative samples was applied to Whatman 3MM filter paper. Filter papers were air-dried overnight and stored at room temperature in individual plastic bags with desiccant.
DNA extraction and speciation
Molecular analysis was conducted at the Howard Hughes Medical Institute/University of Maryland School of Medicine in Baltimore, MD, USA. DNA was extracted from the blood-impregnated filter papers using the QIAmp 96 DNA kit (Qiagen, Valencia, CA, USA). Plasmodium species (P. vivax and P. falciparum) were confirmed by nested PCR amplification of the small subunit ribosomal ribonucleic acid (ssrRNA) genes by using the primers and thermal cycler conditions as previously described [41]. PCR products were visualized by 2–2.5% agarose gel electrophoresis using the Bio-Rad gel doc system. Extracted DNA was stored at −20°C for further analysis.
Pyrosequencing of pfdhfr, pfdhps, pfcrt, and pfmdr1 codons
PCR for pyrosequencing of pfdhfr codons C50R, N51I, C59R, S108N and I164L and pfdhps codons S436A, A437G, K540E, A581G and A613T/S was conducted using previously described cycling conditions [42] with a few modifications. Nested PCR reactions were prepared in 25 μl PCR reaction volume, contained 1 μl DNA template, 1× PCR buffer (Qiagen, Valencia, CA, USA), 0.2 mM dNTPs (Invitrogen) and 0.05 units HotStar Taq DNA polymerase (Qiagen, Valencia, CA, USA). The substitution of HotStar Taq required an initial incubation step of at 95°C for 15 min for all PCR reactions. PCR was performed busing a BioRad tetrad Thermal cycler (Bio-Rad, Hercules, CA). PCR primers and cycling conditions for pyrosequencing of pfcrt codons 72–76 and pfmdr1 codons N86Y, Y184F, S1034C, N1042D and D1246Y are described on the investigator’s website [43].
Pyrosequencing for all genes was performed using PyroMark® Q96 MD pyrosequencer, using the protocol previously described [42]. Between 2 and 7 μl of secondary PCR product or sequence-specific amplified positive control DNA provided by Malaria Research Reagent Resource (MR4) (Manassas, Virginia, USA) was added to each pyrosequencing reaction, depending on the on the PCR product yield. Single nucleotide polymorphisms (SNPs) were called using PyroMark® Q96 MD pyrosequencing software version 1.2 (Qiagen) in allele quantification mode (AQ) for all SNPs except for pfcrt 72– 76 and pfmdr1 N86Y, which were analysed in sequence analysis mode (SQA). Single peak signals of at least 30 RLU (relative luminescence units) were considered suitable for allele quantification. Pyrosequencing was repeated with an adjusted amount of PCR product for samples failing to produce a SNP call because of too much DNA (saturating the pyrogram) or insufficient DNA (producing a weak signal).
SNP calls obtained by pyrosequencing were adjusted using standard curve equations derived from pyrosequencing of mixtures of control DNA strains with known proportions for each allele. To account for machine error, the lower cutoff for inclusion of minor alleles in a mixed infection was set at 10%. pfcr t K76T and pfmdr 1 N86Y could not be absolutely quantified using pyrosequencing so a standard curve could not be applied. The minor allele frequency cutoff for these two codons was set at 20% to account for both machine error and lack of adjustment by standard curves.
Isolates were considered to be ‘pure’ mutants if only the resistance-conferring codon was observed, and ‘mixed’ if both the wild-type and mutant allele were detected. Similarly, haplotypes (combinations of polymorphisms) were considered ‘pure’ mutants if wild-type alleles were not detected in any of the SNP positions, and mixed if both wild-type and mutant alleles were detected in at least one SNP position.
pfmdr1 copy number variation
pfmdr1 copy number was assessed by TaqMan real-time PCR (ABI sequence detector 7700; Applied Biosystems, Warrington, UK) using previously described primers, probes, and reaction conditions [33]. Every qPCR run contained genomic DNA extracted from P. falciparum strains of 3D7, Dd2 and K1 with pfmdr1 copy numbers, of 1, 2, and 3 respectively. Each sample in this assay was run in triplicate. Assays were repeated if one of the following three results was obtained: ∆∆Ct spread > 1.5; Ct values > 35; or copy number estimates between 1.3 and 1.6 [33].
Microsatellite genotyping
Six microsatellites, located 2.8, 4.3, 10.8, and 29.3 kb upstream of pfcrt and 0.6 and 10.4 downstream of pfcrt on chromosome 7, were used for genotyping. These microsatellites were amplified using heminested PCR and fluorescently labeled primers [44]. Specifics of PCR reactions, thermal cycling conditions, and fragment size calling were carried out as described by Laufer et al.[45]. In addition to samples collected in this study, four P. falciparum DNA samples collected between 2005 and 2007 in a previous study in Pakistan [15] and four collected in Dagua Province in Papua New Guinea in 2010 were genotyped at all six microsatellite loci to assess shared ancestry of the pfcrt locus.
Results
A total of 801 microscopy-positive samples was collected from 14 sites. Of these isolates, 128 samples were identified by PCR as P. falciparum and an additional 43 samples contained both P. falciparum and P. vivax infections (Table 1) for a total of 171 samples containing P. falciparum. Sixty three percent (108 samples) were collected from male subjects and 37% (63 samples) were collected from females. Subject age ranged from three months to 75 years, with a median of 24 years.
Pyrosequencing of pfdhfr and pfdhps genes harbouring mutations conferring drug resistance was carried out in 171 PCR-positive P. falciparum isolates. The pfdhfr triple mutant N51I + C59R + S108N was detected in 7 samples, all carrying a mixture of wild-type and mutant alleles (Table 2). The pfdhfr double mutant C59R + S108N was found in 148 samples (87%) (Table 2). The pfdhps mutant codon A437G was observed in 61 samples (51.5%), with few mixed infections. pfdhps K540E was found in six samples, and mixed K/E infections were found in 131 samples (77%). pfdhps A581G was observed in five samples while 66 samples (39%) had the mixed A/G genotype. pfdhfr/pfdhps C59R + S108N + A437G was found in 88 isolates (51%).
The distribution of common pfdhfr and pfdhps haplotypes (combinations of polymorphisms) by region is shown in Table 3. Prevalence of pfdhfr double mutant C59R + S108N ranged from 72% in Punjab (in 31 of 43 samples) to 95% in Khyber Pakhtunkhwa (in 87 of 91 samples). pfdhfr C59R + S108N + pfdhps A437G was present in 44-58% of samples collected in Punjab, Khyber Pakhtunkhwa, and Balochistan (sample sizes of 43, 91, and 33, respectively). Only two P. falciparum samples each was collected from Islamabad and Sindh, and all four carried pfdhfr C59R + S108N + pfdhps A437G.
All 171 samples analysed were amplified successfully for pfcrt. DNA pyrosequencing of the secondary PCR products from these samples confirmed that all of the amplified samples carried codon K76T (nucleotide sequence ACA). All samples were of the SVMNT haplotype for pfcrt 72–76 (Table 2).
Pfmdr1 N86Y pure mutants were identified in six (4%) of the tested isolates, 28 (16%) were identified as mixed N/Y infections (Table 2). Forty-three (25%) isolates carried the mixed Y/F codon at position Y184F of the pfmdr1 gene and the remaining samples carried the wild-type Y codon. One hundred and sixty-five (96%) isolates carried the mixed N/D polymorphism at position N1042D. All samples were wild-type for codons S1034C and D1246Y. Real time PCR to determine copy number variation in pfmdr1 gene revealed that all isolates harboured a single copy of the pfmdr1 gene.
All six microsatellite loci amplified successfully in 160 of 171 P.falciparum samples. Three isolates were mixed-strain infections with multiple alleles at each locus and were excluded from further analysis. Three haplotypes were observed in the remaining sample set of 157: the predominant haplotype, found in 137 samples, and two haplotypes in 12 and eight samples, respectively, each differing from the predominant haplotype at one microsatellite locus (Table 4). These pfcrt flanking microsatellite haplotypes were identical or nearly identical (differing in 1 locus) to those found in isolates with the SVMNT haplotype from Pakistan [15] and Papua New Guinea (Table 4).
Discussion
In this study, polymorphisms in the pfdhfr, pfdhps, pfcrt and pfmdr1 genes were investigated in P. falciparum samples collected throughout Pakistan to determine the current extent of resistance to chloroquine and antifolates, both of which are still used as part of first-line treatment of malaria in Pakistan.
The complete saturation of pfcrt K76T in all parts of the country indicates that chloroquine-resistant P. falciparum is fixed in Pakistan. These findings corroborate previous molecular studies reporting near fixation of chloroquine resistance [15, 37] and clinical studies reporting high treatment failure of chloroquine [2, 7, 8]. Chloroquine resistance has also been documented in several neighbouring countries, including Iran, India, China and Afghanistan [46–50]. Although chloroquine is recommended for P. vivax infections only, P. falciparum infections are often exposed to this treatment as well. Presumptive diagnosis based on clinical grounds, improper diagnosis, lack of diagnostic facilities, and empirical treatment are common practices in resource-limited countries such as Pakistan [51, 52], and these factors likely result in the continued mistreatment of P. falciparum infections with chloroquine. Such ineffective treatment of chloroquine-resistant P. falciparum with chloroquine can lead to increased morbidity and mortality if infections do not respond to the drug. Use of chloroquine to treat P. falciparum has also been postulated as contributing to the rapid growth of falciparum malaria in Pakistan [2], as infections may not clear and continue to circulate in the population unchecked.
Typing of microsatellites surrounding the pfcrt locus indicates that a selective sweep of chloroquine-resistant P. falciparum occurred in Pakistan, and that the SVMNT allele in Pakistan shares its ancestry with parasites in Papua New Guinea. Previous studies have shown that the SVMNT allele of the pfcrt locus appears to have swept through Papua New Guinea [14], multiple parts of India [16, 46] and a small number of sites Pakistan [15]. Taken together with the findings in this study, these results suggest that the spread of SVMNT may have been a single sweep event through a large region encompassing parts of the South Pacific and South Asia. The possibility that such a large sweep could occur again in the context of artemisinin resistance in this region should be carefully considered.
The current first-line treatment for confirmed P. falciparum malaria in Pakistan is artesunate + SP. The pfdhfr C59R + S108N double mutant was the most frequent pyrimethamine-resistant haplotype found, as has been shown in previous studies in Pakistan [1, 37], India [47, 48], Iran [38, 49], and Sri Lanka [50]. The majority of samples also carried a mutation at position A437G and a number of samples harboured mixed infections at highly resistant codons K540E and A581G in the pfdhps gene. Although the clinically significant pfdhfr triple mutation was rare (and present only in mixed wild-type/mutant infections) and pfdhfr/pfdhps quintuple mutants were not detected, the presence of multilocus resistance haplotypes and the presence of highly resistant sulphadoxine alleles indicate that clinically relevant SP resistance could arise or be already present. A recent clinical trial in India reported artesunate + SP efficacy dipping to near the WHO-established efficacy threshold of 90%, likely attributable to the presence of multiple pfdhfr and pfdhps mutations [53].
All samples harboured a single copy of the pfmdr1 gene, suggesting that artesunate, lumefantrine, and mefloquine are likely to have high efficacy in Pakistan, and that the latter two could be considered as alternate ACT partner drugs if the prevalence of SP resistance markers rises. Presence of point mutations at position N86Y, Y184F and N1042D of pfmdr1 indicate that continued molecular monitoring of this gene may provide informative insights on its association with artemisinin and partner drugs in this region.
Conclusions
The fixation of chloroquine resistance marker pfcrt K76T in Pakistan suggests that, in regions with known high levels of P. falciparum, improved species diagnosis and appropriate treatment with ACT are critical to ensure that malaria patients receive effective care. The presence of multiple resistance alleles in pfdhfr and pfdhps demonstrates the utility of molecular surveillance to monitor the efficacy of artesunate + SP in Pakistan. Based on these findings, molecular and therapeutic efficacy studies to assess the continued efficacy of artesunate + SP are warranted.
References
Ghanchi NK, Ursing J, Beg MA, Veiga MI, Jafri S, Martensson A: Prevalence of resistance associated polymorphisms in Plasmodium falciparum field isolates from southern Pakistan. Malar J. 2011, 10: 18-10.1186/1475-2875-10-18.
Shah I, Rowland M, Mehmood P, Mujahid C, Razique F, Hewitt S, Durrani N: Chloroquine resistance in Pakistan and the upsurge of falciparum malaria in Pakistani and Afghan refugee populations. Ann Trop Med Parasitol. 1997, 91: 591-602. 10.1080/00034989760680.
Bouma MJ, Dye C, van der Kaay HJ: Falciparum malaria and climate change in the northwest frontier province of Pakistan. Am J Trop Med Hyg. 1996, 55: 131-137.
Rab MA, Freeman TW, Durrani N, de Poerck D, Rowland MW: Resistance of Plasmodium falciparum malaria to chloroquine is widespread in eastern Afghanistan. Ann Trop Med Parasitol. 2001, 95: 41-46. 10.1080/00034980020035906.
Durrani AB, Durrani IU, Abbas N, Jabeen M: Epidemiology of cerebral malaria and its mortality. J Pak Med Assoc. 1997, 47: 213-215.
WHO: World malaria report 2011. 2011, Geneva: World Health Organization
Rowland M, Durrani N, Hewitt S, Sondorp E: Resistance of falciparum malaria to chloroquine and sulfadoxine-pyrimethamine in Afghan refugee settlements in western Pakistan: surveys by the general health services using a simplified in vivo test. Trop Med Int Health. 1997, 2: 1049-1056. 10.1046/j.1365-3156.1997.d01-185.x.
Howard N, Durrani N, Sanda S, Beshir K, Hallett R, Rowland M: Clinical trial of extended-dose chloroquine for treatment of resistant falciparum malaria among Afghan refugees in Pakistan. Malar J. 2011, 10: 171-10.1186/1475-2875-10-171.
WHO: World malaria report 2008. 2008, Geneva: World Health Organization
WHO: Division of communicable disease control newsletter. 2005, Geneva: World Health Organization
Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE: Mutations in the P. falciparum digestive vacuole transmembrane protein Pfcrt and evidence for their role in chloroquine resistance. Mol Cell. 2000, 6: 861-871. 10.1016/S1097-2765(05)00077-8.
Wellems TE, Plowe CV: Chloroquine-resistant malaria. J Infect Dis. 2001, 184: 770-776. 10.1086/322858.
Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV: A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001, 344: 257-263. 10.1056/NEJM200101253440403.
Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ: Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002, 418: 320-323. 10.1038/nature00813.
Rawasia WF, Sridaran S, Patel JC, Abdallah J, Ghanchi NK, Barnwell JW, Escalante AA, Udhayakumar V, Beg MA: Genetic backgrounds of the Plasmodium falciparum chloroquine resistant transporter (pfcrt) alleles in Pakistan. Infect Genet Evol. 2012, 12: 278-281. 10.1016/j.meegid.2011.11.008.
Mixson-Hayden T, Jain V, McCollum AM, Poe A, Nagpal AC, Dash AP, Stiles JK, Udhayakumar V, Singh N: Evidence of selective sweeps in genes conferring resistance to chloroquine and pyrimethamine in Plasmodium falciparum isolates in India. Antimicrob Agents Chemother. 2010, 54: 997-1006. 10.1128/AAC.00846-09.
DaRe JT, Mehlotra RK, Michon P, Mueller I, Reeder J, Sharma YD, Stoneking M, Zimmerman PA: Microsatellite polymorphism within pfcrt provides evidence of continuing evolution of chloroquine-resistant alleles in Papua New Guinea. Malar J. 2007, 6: 34-10.1186/1475-2875-6-34.
Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ: Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988, 85: 9109-9113. 10.1073/pnas.85.23.9109.
Peterson DS, Walliker D, Wellems TE: Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988, 85: 9114-9118. 10.1073/pnas.85.23.9114.
Peterson DS, Milhous WK, Wellems TE: Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci U S A. 1990, 87: 3018-3022. 10.1073/pnas.87.8.3018.
Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE: Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995, 52: 565-568.
Plowe CV, Kublin JG, Doumbo OK: P. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Resist Updat. 1998, 1: 389-396. 10.1016/S1368-7646(98)80014-9.
Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RA, Rogerson SJ, Lescano AG, Molyneux ME, Winstanley PA, Chimpeni P, Taylor TE, Plowe CV: Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002, 185: 380-388. 10.1086/338566.
Brooks DR, Wang P, Read M, Watkins WM, Sims PF, Hyde JE: Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. 1994, 224: 397-405. 10.1111/j.1432-1033.1994.00397.x.
Foote SJ, Galatis D, Cowman AF: Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc Natl Acad Sci USA. 1990, 87: 3014-3017. 10.1073/pnas.87.8.3014.
Triglia T, Cowman AF: Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1994, 91: 7149-7153. 10.1073/pnas.91.15.7149.
Triglia T, Menting JG, Wilson C, Cowman AF: Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997, 94: 13944-13949. 10.1073/pnas.94.25.13944.
Wang P, Read M, Sims PF, Hyde JE: Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol. 1997, 23: 979-986. 10.1046/j.1365-2958.1997.2821646.x.
Happi CT, Gbotosho GO, Folarin OA, Akinboye DO, Yusuf BO, Ebong OO, Sowunmi A, Kyle DE, Milhous W, Wirth DF, Oduola AM: Polymorphisms in Plasmodium falciparum dhfr and dhps genes and age related in vivo sulfadoxine-pyrimethamine resistance in malaria-infected patients from Nigeria. Acta Trop. 2005, 95: 183-193. 10.1016/j.actatropica.2005.06.015.
Picot S, Olliaro P, de Monbrison F, Bienvenu AL, Price RN, Ringwald P: A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J. 2009, 8: 89-10.1186/1475-2875-8-89.
Staedke SG, Kamya MR, Dorsey G, Gasasira A, Ndeezi G, Charlebois ED, Rosenthal PJ: Amodiaquine, sulfadoxine/pyrimethamine, and combination therapy for treatment of uncomplicated falciparum malaria in Kampala, Uganda: a randomised trial. Lancet. 2001, 358: 368-374. 10.1016/S0140-6736(01)05557-X.
Lim P, Wongsrichanalai C, Chim P, Khim N, Kim S, Chy S, Sem R, Nhem S, Yi P, Duong S, Bouth DM, Genton B, Beck HP, Gobert JG, Rogers WO, Coppee JY, Fandeur T, Mercereau-Puijalon O, Ringwald P, Le BJ, Ariey F: Decreased in vitro susceptibility of Plasmodium falciparum isolates to artesunate, mefloquine, chloroquine, and quinine in Cambodia from 2001 to 2007. Antimicrob Agents Chemother. 2010, 54: 2135-2142. 10.1128/AAC.01304-09.
Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S: Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004, 364: 438-447. 10.1016/S0140-6736(04)16767-6.
Price RN, Uhlemann AC, van VM, Brockman A, Hutagalung R, Nair S, Nash D, Singhasivanon P, Anderson TJ, Krishna S, White NJ, Nosten F: Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis. 2006, 42: 1570-1577. 10.1086/503423.
Alker AP, Lim P, Sem R, Shah NK, Yi P, Bouth DM, Tsuyuoka R, Maguire JD, Fandeur T, Ariey F, Wongsrichanalai C, Meshnick SR: Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am J Trop Med Hyg. 2007, 76: 641-647.
Petersen I, Eastman R, Lanzer M: Drug-resistant malaria: molecular mechanisms and implications for public health. FEBS Lett. 2011, 585: 1551-1562. 10.1016/j.febslet.2011.04.042.
Khatoon L, Baliraine FN, Bonizzoni M, Malik SA, Yan G: Prevalence of antimalarial drug resistance mutations in Plasmodium vivax and P. falciparum from a malaria-endemic area of Pakistan. Am J Trop Med Hyg. 2009, 81: 525-528.
Zakeri S, Gil JP, Bereckzy S, Djadid ND, Bjorkman A: High prevalence of double Plasmodium falciparum dhfr mutations at codons 108 and 59 in the Sistan-Baluchistan province, Iran. J Infect Dis. 2003, 187: 1828-1829. 10.1086/375250.
Nawaz S: FATA—A Most Dangerous Place. 2009, Washington, D.C
Asif SA: Departmental audit of malaria control programme 2001–2005 north west frontier province (NWFP). J Ayub Med Coll Abbottabad. 2008, 20: 98-102.
Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN: Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993, 58: 283-292. 10.1016/0166-6851(93)90050-8.
Zhou Z, Poe AC, Limor J, Grady KK, Goldman I, McCollum AM, Escalante AA, Barnwell JW, Udhayakumar V: Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum. J Clin Microbiol. 2006, 44: 3900-3910. 10.1128/JCM.01209-06.
CVD Malaria Group: Protocols. [http://medschool.umaryland.edu/malaria/protocols.asp]
Nash D, Nair S, Mayxay M, Newton PN, Guthmann JP, Nosten F, Anderson TJ: Selection strength and hitchhiking around two anti-malarial resistance genes. Proc Biol Sci. 2005, 272: 1153-1161. 10.1098/rspb.2004.3026.
Laufer MK, Takala-Harrison S, Dzinjalamala FK, Stine OC, Taylor TE, Plowe CV: Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J Infect Dis. 2010, 202: 801-808. 10.1086/655659.
Lumb V, Madan R, Das MK, Rawat V, Dev V, Khan W, Khan H, Sharma YD: Differential genetic hitchhiking around mutant pfcrt alleles in the Indian Plasmodium falciparum population. J Antimicrob Chemother. 2012, 67: 600-608. 10.1093/jac/dkr532.
Ahmed A, Bararia D, Vinayak S, Yameen M, Biswas S, Dev V, Kumar A, Ansari MA, Sharma YD: Plasmodium falciparum isolates in India exhibit a progressive increase in mutations associated with sulfadoxine-pyrimethamine resistance. Antimicrob Agents Chemother. 2004, 48: 879-889. 10.1128/AAC.48.3.879-889.2004.
Biswas S, Escalante A, Chaiyaroj S, Angkasekwinai P, Lal AA: Prevalence of point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum isolates from India and Thailand: a molecular epidemiologic study. Trop Med Int Health. 2000, 5: 737-743. 10.1046/j.1365-3156.2000.00632.x.
Zakeri S, Farahani MS, Afsharpad M, Salehi M, Raeisi A, Djadid ND: High prevalence of the 437G mutation associated with sulfadoxine resistance among Plasmodium falciparum clinical isolates from Iran, three years after the introduction of sulfadoxine-pyrimethamine. Int J Infect Dis. 2010, 14 (Suppl 3): e123-e128.
Hapuarachchi HC, Dayanath MY, Bandara KB, Abeysundara S, Abeyewickreme W, de Silva NR, Hunt SY, Sibley CH: Point mutations in the dihydrofolate reductase and dihydropteroate synthase genes of Plasmodium falciparum and resistance to sulfadoxine-pyrimethamine in Sri Lanka. Am J Trop Med Hyg. 2006, 74: 198-204.
Parikh R, Amole I, Tarpley M, Gbadero D, Davidson M, Vermund SH: Cost comparison of microscopy vs. empiric treatment for malaria in Southwestern Nigeria: a prospective study. Malar J. 2010, 9: 371-10.1186/1475-2875-9-371.
Kakar Q, Khan MA, Bile KM: Malaria control in Pakistan: new tools at hand but challenging epidemiological realities. East Mediterr Health J. 2010, 16 (Suppl): S54-S60.
Saha P, Guha SK, Das S, Mullick S, Ganguly S, Biswas A, Bera DK, Chattopadhyay G, Das M, Kundu PK, Ray K, Maji AK: Comparative efficacies of artemisinin combination therapies in Plasmodium falciparum malaria and polymorphism of pfATPase6, pfcrt, pfdhfr, and pfdhps genes in tea gardens of Jalpaiguri District, India. Antimicrob Agents Chemother. 2012, 56: 2511-2517. 10.1128/AAC.05388-11.
Acknowledgements
The authors thank the study participants for their involvement in the study. The authors also acknowledge Matthew Adams, Malathi Vadla and Sudhaunshu Joshi at the University of Maryland for their technical assistance and Muhammad Shahid, Ismail Jalil, Ifthikhar Ahmed, Adeel Riaz and Noor Rehman in Pakistan for help in sample collection. The authors would like to thank Venkatachalam Udhayakumar at the Centers for Disease Control and Prevention for generously providing control DNA from Pakistan for microsatellite validation. This work was supported by a grant from the Higher Education Commission of Pakistan in support of AAK’s PhD studies at Quaid-i-Azam University, Islamabad, Pakistan, and was supported in part by the Howard Hughes Medical Institute at the University of Maryland School of Medicine, Baltimore, MD, USA.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AAK designed study, carried out the laboratory experiments, and performed data analysis. AAK and MV drafted the manuscript. CGJ, MA and EMA contributed to laboratory experiments and data analysis. MFN and FN participated in sample data collection and microscopy. FH and TM contributed to sample collection and contributed advice for microsatellite data analysis. MV, SAM and CVP provided guidance and coordination for study design, genotyping, and data analyses, and edited and revised the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Khattak, A.A., Venkatesan, M., Jacob, C.G. et al. A comprehensive survey of polymorphisms conferring anti-malarial resistance in Plasmodium falciparum across Pakistan. Malar J 12, 300 (2013). https://doi.org/10.1186/1475-2875-12-300
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-12-300