Abstract
Background
Plasmodium vivax is a significant contributor to the global malaria burden, and a vaccine targeting vivax malaria is urgently needed. An understanding of the targets of functional immune responses during the course of natural infection will aid in the development of a vaccine. Antibodies play a key role in this process, with responses against particular epitopes leading to immune selection pressure on these epitopes. A number of techniques exist to estimate levels of immune selection pressure on particular epitopes, with a sliding window analysis often used to determine particular regions likely to be under immune pressure. However, such analysis neglects protein three-dimensional structural information. With this in mind, a newly developed tool, BioStructMap, was applied to two key antigens from Plasmodium vivax: PvAMA1 and PvDBP Region II. This tool incorporates structural information into tests of selection pressure.
Results
Sequences from a number of populations were analysed, examining spatially-derived nucleotide diversity and Tajima’s D over protein structures for PvAMA1 and PvDBP. Structural patterns of nucleotide diversity were similar across all populations examined, with Domain I of PvAMA1 having the highest nucleotide diversity and displaying significant signatures of immune selection pressure (Tajima’s D > 0). Nucleotide diversity for PvDBP was highest bordering the dimerization and DARC-binding interface, although there was less evidence of immune selection pressure on PvDBP compared with PvAMA1. This study supports previous work that has identified Domain I as the main target of immune-mediated selection pressure for PvAMA1, and also supports studies that have identified functional epitopes within PvDBP Region II.
Conclusions
The BioStructMap tool was applied to leading vaccine candidates from P. vivax, to examine structural patterns of selection and diversity across a number of geographic populations. There were striking similarities in structural patterns of diversity across multiple populations. Furthermore, whilst regions of high diversity tended to surround conserved binding interfaces, a number of protein regions with very low diversity were also identified, and these may be useful targets for further vaccine development, given previous evidence of functional antibody responses against these regions.
Similar content being viewed by others
Background
Plasmodium vivax infected an estimated 8.55 million people in 2016 and is a significant contributor to global malaria morbidity, with the majority of P. vivax cases occurring within South-East Asia [1]. There remains a significant need for a vaccine against P. vivax, and an understanding of the targets of natural immune responses following P. vivax infection is likely to aid such an effort. A key challenge in vaccine development is the identification of specific antigens and epitopes that are targets of protective antibody responses. It is possible to use population genetic data to identify regions of proteins that are under immune-mediated selection pressure, which gives rise to balancing selection within that protein region. Tajima’s D is one test statistic that is often used to identify departures from a neutral model of selection, and has been applied to malaria genes both on a per-gene basis [2, 3], or as a sliding window analysis along a gene [4,5,6,7]. A number of studies have previously examined P. vivax proteins such as apical membrane antigen 1 (PvAMA1) and Duffy-binding protein (PvDBP) for evidence of immune selection pressure using these approaches [4, 8,9,10,11]. However, because a sliding window analysis is typically performed over the linear gene sequence, it does not take into account the impact of the three-dimensional (3D) structural constraints of the protein in the calculation of selection pressures. A new method that allows incorporation of protein structural information into tests for selection pressure has recently been described [12], and has been applied here to two leading P. vivax vaccine candidates: PvAMA1 and PvDBP.
AMA1 is a type I transmembrane protein present in all Plasmodium species [13]. It is localized to the parasite micronemes, and is released onto the surface of the merozoite prior to invasion of red blood cells [14]. AMA1 binds to RON2 during the formation of the tight junction between parasite and host–cell membranes [15, 16] and is a target of protective immune responses [17,18,19,20]. The ectodomain of Plasmodium AMA1 proteins is divided into three domains, termed Domains I (DI), II (DII) and III (DIII) (Additional file 1) [21]. DI is considered to be the most polymorphic, and is also the site of RON2 binding [22]. RON2 binds a conserved hydrophobic cleft that is surrounded by a number of highly polymorphic regions, the most notable being the C1L loop, a surface exposed loop with high variability that is suggested to define strain-specificity in anti-AMA1 responses in Plasmodium falciparum infection [23]. While DI is generally considered to be the most important for functional antibody responses, there is evidence that DII and DIII may also be targets of functional antibody responses in P. falciparum [8, 24]. A number of studies have investigated selection pressures on PvAMA1. Evidence for balancing selection within Domain I has been observed in a Venezuelan population [9], in a number of Papua New Guinean populations (Madang and Wosera, Madang and East Sepik) [4, 8], an Iranian population [10] and in a Peruvian population [25]. In contrast, two other studies examining PvAMA1 sequences in isolates from Korea [26] and Myanmar [27] did not find any evidence of balancing selection, but instead observed evidence of recent population bottleneck and expansion in those populations.
PvDBP is an important micronemal protein that binds to the Duffy antigen/receptor for chemokines (DARC) on human reticulocytes during invasion [28, 29]. Whilst there is evidence that PvDBP is not absolutely essential for invasion of reticulocytes [30,31,32], Duffy-negative individuals are largely resistant to P. vivax infection, and hence PvDBP makes an attractive vaccine target [33]. PvDBP is part of the erythrocyte-binding like (EBL) family of proteins, which include EBA175, EBA181 and EBA140 in P. falciparum [34]. PvDBP is the sole EBL family protein in P. vivax [35]. EBL family proteins are composed of a number of distinct domains, with Region II (RII) being a cysteine-rich Duffy-binding like (DBL) domain that is involved in binding to erythrocytes. EBL family proteins each recognize a different receptor via their respective DBL domains [29, 36,37,38]; PvDBP binds to DARC via its DBL domain (RII) [39]. During this process two PvDBP molecules form a dimer around two DARC molecules [39, 40]. RII of PvDBP has been divided into a number of subdomains (subdomains 1–3) [41] (Additional file 2), and it is subdomain 2 that contains both the dimer interface and DARC binding residues [39]. Immune responses against PvDBP have been associated with protection from clinical malaria in naturally exposed cohorts [42, 43] whilst antibodies against PvDBP RII epitopes have been found to inhibit both attachment of PvDBP RII to erythrocytes [44] and in vitro invasion of erythrocytes [45]. With regards to immune selection pressure on PvDBP, a study of 100 Sri Lankan isolates found no evidence of significant selection pressure on this region using Tajima’s D, dN/dS or Fu and Li’s D and F statistics [11]. Another study examining genetic diversity of PvDBP RII across multiple populations showed a significantly positive value of dN/dS in this region, suggesting that this region may be under immune selection pressure [25].
In this study, selection pressures on PvAMA1 and PvDBP Region II were examined in the context of protein structure, using a newly developed tool called BioStructMap [12]. BioStructMap enables the application of a 3D sliding window over a protein structure. This allows incorporation of protein structural information into tests such as Tajima’s D or nucleotide diversity that are traditionally performed as a linear 2D sliding window over a protein or nucleotide sequence. A previous study identified a discontinuous region of PfAMA1 bordering DII and DIII that had a strong signature of balancing selection when considering spatially derived Tajima’s D [12]. Given that other studies have identified DI of PvAMA1, rather than DII or DIII, as being under balancing selection, it was considered that incorporation of protein structural information might yield additional insights into other regions under immune selection pressure. Genomic sequences from a number of populations were analysed, and spatially-derived nucleotide diversity and Tajima’s D were examined using protein structural information for PvAMA1 and PvDBP Region II. Structural patterns of nucleotide diversity were similar across all populations examined, with Domain I of PvAMA1 having the highest nucleotide diversity and displaying significant signatures of balancing selection (Tajima’s D > 0). Nucleotide diversity for PvDBP was highest bordering the dimerization and DARC-binding interface, although there was less evidence of immune selection pressure on this antigen.
Methods
Data sources
Reference sequences for Sal-1 PvDBP (PVX_110810) and PvAMA1 (PVX_092275) were obtained from PlasmoDB, v34 (http://www.plasmoDB.org) [46]. Genomic sequences from field isolates were extracted from GenBank, restricted to P. vivax, and with the condition that sequences had to cover > 95% of the structured domains examined here (i.e. partial fragments from these regions were excluded). Sequences from non-human hosts were excluded, as were sequences without a known geographic location described either in the associated literature or clearly annotated in the sequence record. Single isolate populations were also excluded. For PvDBP, bases 766–1659 were used, while for PvAMA1 bases 121–1422 were used.
Sequence polymorphism analysis
The DendroPy Python library [47] was used to calculate Tajima’s D, Watterson’s Theta, mean pairwise differences and nucleotide diversity. Haplotype diversity was calculated using:
where xi is the relative frequency of the ith haplotype and n is the number of samples.
Normalized Shannon entropy [48] for each position in the protein sequence was calculated using:
where S is the normalized Shannon entropy and p i is the frequency at that position of the ith amino acid in the standard 20 amino acid alphabet. The normalized Shannon entropy is a measure of sequence diversity, and takes values between 0 and 1, where 0 indicates perfect sequence conservation at that site, and 1 indicates an even distribution of all possible amino acids at that site.
Phylogenetic analysis
Sequences from all isolates were aligned using MUSCLE v3.8.31 [49], and alignments manually adjusted to minimize gaps. Maximum likelihood phylogenetic trees were constructed for AMA-1 and DBP sequences using IQ-TREE v1.3.11.1 [50]. The ultrafast bootstrap estimation (UFBoot) [51] was used with 5000 bootstrap replicates and a best fit model was chosen according to the Bayesian Inference Criterion [52]. Phylogenetic trees were visualized with iTOL [53].
Incorporation of structural information into sequence analysis
A Python package, BioStructMap, which allows for the application of a 3D sliding window over a protein structure has been previously described [12]. BioStructMap was used to compute spatially derived Tajima’s D and nucleotide diversity (π) values for both PvAMA1 and PvDBP structures, with a radius of 15 Å for each window. BioStructMap was also used to calculate Normalized Shannon Entropy on a per-residue basis.
Protein structural models
Known PvDBP structures (i.e. 4NUU, 3RRC, 4YFS, 4NUV, 5F3J) all contain a number of unresolved residues, and to ensure complete coverage of DBP Region II in this analysis, a template-based model of P. vivax DBP was generated for use with BioStructMap. This model was created using ModPipe [54], an automated software pipeline that utilizes MODELLER for the generation of comparative protein structure models [55]. The PDB structure 4NUU was used to generate this comparative model. The generated model is accessible via ModBase (https://modbase.compbio.ucsf.edu/; ModPipe model ID f7602e019fac5be4a79c4cca6751b392) [56].
The PvAMA1 model used has been previously described [4], and uses a chimeric template to generate a structural model of PvAMA1.
Comparing patterns of selection and diversity between populations
To compare structural patterns of nucleotide diversity and Tajima’s D between all populations considered in this study, Spearman’s rank correlation coefficient was computed for both PvAMA1 and PvDBP residue data between each pair of populations. Residues with missing data in one or both populations (i.e. Tajima’s D was undefined) were excluded from analysis for that pair of populations.
Data analysis, statistics and other software used
The majority of data analysis was performed using the Anaconda distribution of Python 3.5. Plotting was performed with the Python Matplotlib package, version 1.5.1 [57]. Statistical analysis was performed using SciPy [58]. Protein structures were visualized using PyMol [59].
Results
Population structure of PvAMA1 and PvDBP sequences
This study aimed to examine selection pressures on key structured domains of PvDBP and PvAMA1. Genomic sequences for each antigen were extracted from GenBank, with a total of 505 PvAMA1 sequences and 243 PvDBP sequences obtained, belonging to 10 and 12 distinct populations, respectively. Sequences which did not cover at least 95% of the structured domains examined (PvAMA1, PVX_092275: nucleotides 121–1422; PvDBP, PVX_110810: nucleotides 766–1659) were excluded from analysis, as were sequences from single-isolate populations or non-human hosts. Maximum likelihood phylogenetic trees were constructed for both PvAMA1 and PvDBP (Fig. 1) using aligned sequences from all populations. Some populations were generally contained on their own branch (e.g. South Korea for PvAMA1, Mexico and Papua New Guinea for PvDBP), while other branches contained a mix of populations. This intermixing was particularly evident for populations that are geographically close, such as Thailand, Myanmar and Papua New Guinea for PvAMA1.
Traditional measures of selection pressure and diversity for PvAMA1 and PvDBP
Key population parameters for PvAMA1 and PvDBP sequences were computed, as well as several measures of diversity and selection (Tables 1, 2). A total of 259 haplotypes were observed for PvAMA1 (haplotype diversity = 0.99), while 84 haplotypes were observed for PvDBP RII (haplotype diversity = 0.96).
For PvAMA1, nucleotide diversity (π) and mean number of pairwise differences (k) were highest in the three Thai populations examined, with a maximum π of 10.09 × 10−3 in the 2007 Tak Province, Thailand samples. Nucleotide diversity was lowest in the South Korean population (π = 5.90 × 10−3), which may be explained by a recent bottleneck in this population, limiting overall diversity; this is supported by the negative Tajima’s D value observed for PvAMA1 in this South Korean population (D = − 1.20). Tajima’s D is often used to identify regions under balancing selection (Tajima’s D > 0), which can be the result of immune selection pressure, and it is noted that for PvAMA1, only the Venezuelan population had a significantly positive Tajima’s D value (D = 2.12, p < 0.05), as per the confidence limits outlined in Tajima [60], although these limits may be overly conservative [61]. Most other populations also had a positive Tajima’s D value, such as Papua New Guinea (Madang: D = 1.59; East Sepik: D = 1.09) and Thailand (Chantaburi: D = 1.22), with the sole exception of South Korea (discussed above).
When examining PvDBP, nucleotide diversity was highest in South Korea (π = 10.53 × 10−3), Bangkok, Thailand (π = 10.37 × 10−3) and Myanmar (π = 10.17 × 10−3). Interestingly, the South Korean population had a negative Tajima’s D value that was statistically significant, suggestive of a recent population bottleneck and expansion. This population also had the highest number of polymorphic sites (58 non-synonymous). This high variability is probably due to 2 or 3 divergent isolates (Fig. 1b), and haplotype diversity is relatively low for this population (Hd = 0.85). Tajima’s D values for PvDBP were mostly close to zero, with the highest value observed for a Colombian population (D = 1.72).
These observations agree with previous studies that have generally observed greater signatures of immune selection pressure on PvAMA1 as compared to PvDBP [25].
Diversity and selection on the PvAMA1 structure
We then examined selection pressures and polymorphisms in the context of protein 3D structure using BioStructMap, a tool that allows for incorporation of protein structural information into a sliding window analysis. To quantify nucleotide diversity over the PvAMA1 structure both as a spatially averaged value and a per-residue value, nucleotide diversity was calculated using a 3D sliding window (Fig. 2a) and per-residue normalized Shannon entropy (Fig. 2b) using sequences from all populations. Normalized Shannon entropy is a measure of sequence diversity, taking values between 0 and 1, where 0 indicated perfect sequence conservation at that position, while 1 indicates an even distribution of all possible amino acids at that position. For PvAMA1, nucleotide diversity was highest in DI on one side of the RON2 binding cleft (Fig. 2a, c). There was limited diversity within DII, and very little observed in DIII. This pattern of nucleotide diversity appeared to be maintained when examining patterns of nucleotide diversity in individual populations (Additional file 3), with the exception of the South Korean population, in which even DI displays limited diversity. This is possibly due to recent bottleneck and expansion, as suggested by the negative Tajima’s D value for this population. It was also observed that the so-called ‘silent face’ of PvAMA1 had very low diversity in all populations examined, which is in line with a number of other studies that have noted a distinct lack of polymorphisms on this face both in P. falciparum [8, 62,63,64] and P. vivax [4].
Measures of sequence diversity for PvAMA1 and relationship to key binding interfaces. a Spatially-derived nucleotide diversity (π) for PvAMA1 displayed over the modelled PvAMA1 structure. A radius of 15 Å was used for each 3D sliding window. b Normalized Shannon entropy for PvAMA1 residues on a per-site basis, with no spatial averaging performed. Higher entropy values are indicative of greater sequence diversity across all strains at that residue position. c Residues involved in the binding of PvAMA1 to its PvRON2 ligand. Residues within 4 Å of the RON2 peptide are shown in blue on the modelled PvAMA1 structure (as defined by the PDB structure 5NQG). Note that some binding residues are not visible due to the presence of the flexible Domain II loop near the RON2 binding groove
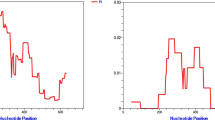
To test for evidence of immune-mediated selection pressure on PvAMA1, Tajima’s D was calculated as a 3D sliding window over the modelled protein structure (referred to as a spatially-derived Tajima’s D). A positive Tajima’s D value provides evidence for balancing selection, which can arise as a result of immune pressure. It is noted that departures from neutrality can also arise as a result of population structure. In particular, sampling of strains across multiple distinct populations can potentially give rise to similar signatures to those of balancing selection. For this reason Tajima’s D values were analysed within distinct geographic and temporal populations. Spatially-derived Tajima’s D was highest in DI for nearly all populations examined, with the exception of Sri Lanka and South Korea (Fig. 3; Additional file 4). As was observed for nucleotide diversity, Tajima’s D was highest on one side of DI, with the other ‘silent-face’ typically having Tajima’s D values close to zero. This is in agreement with several studies, both in P. vivax and P. falciparum, in which polymorphisms on AMA1 are focused on one side of the protein structure, with minimal polymorphic variation on the other face [4, 8, 62,63,64]. It has been hypothesized that this silent face of the protein is not exposed to the immune system during the invasion process due to interaction with other parasite proteins, or is otherwise inaccessible to antibody binding [63].
Diversity and selection on the PvDBP structure
A similar analysis was performed for PvDBP, calculating spatially-derived nucleotide diversity, normalized Shannon entropy and spatially-derived Tajima’s D over the modelled PvDBP structure. Global nucleotide diversity was highest within subdomain 2 of PvDBP, with the most polymorphic residues clustering around the dimerization and DARC binding interface (Fig. 4). Residues that are directly involved in DARC binding and dimerization were nearly all highly conserved. When examining patterns of nucleotide diversity across individual geographic locations (Additional file 5), it is noted that nucleotide diversity within PvDBP RII was universally highest in a region that corresponds to a previously identified inhibitory epitope, termed the DEK epitope [33, 39, 44], which corresponds to residues 338–353 (DEKAQQRRKQWWNESK) of the Sal-1 reference sequence. Correspondingly, the opposite end of the PvDBP protein (subdomain 3) had very low nucleotide diversity across all populations. Spatially derived Tajima’s D values were also calculated for PvDBP, and observed that most populations had Tajima’s D values that were negative or close to zero over most of the structure (Fig. 5; Additional file 6); Tajima’s D values did not reach statistical significance in any population (with the exception of a single residue in samples from Chennai, India) (Additional file 7). However, it was noted that spatially derived Tajima’s D values were generally highest in subdomain 2.
Measures of sequence diversity for PvDBP and relationship to key binding interfaces. a Spatially-derived nucleotide diversity (π) for PvDBP displayed over the modelled PvDBP structure. A radius of 15 Å was used for each 3D sliding window. b Normalized Shannon entropy for PvDBP residues on a per-site basis, with no spatial averaging performed. Higher entropy values are indicative of greater sequence diversity across all strains at that residue position. c Residues involved in the PvDBP dimerization and DARC binding interface. Residues within 4 Å of the corresponding dimer chain or the DARC ligand (as defined by the PDB structure 4NUU) are shown in blue on the modelled PvDBP structure
Amino acid mutations within binding interfaces
While binding interfaces were generally observed to have low nucleotide diversity, there were some polymorphic residues located within these binding interfaces for both PvAMA1 and PvDBP (Additional file 8). There were a total of 9 polymorphic residues within the PvAMA1:RON2 interface, although most of these were present at very low frequencies within the global population. The three most frequent polymorphisms within the PvAMA1:RON2 interface fall within a short stretch from residues 130–133 (N132D, N130K, D133N). The most frequent of these polymorphisms (N132D) is a relatively conservative change from asparagine to aspartic acid, and is the only one of the three residues whose side chains are directly involved in hydrogen bonds with RON2; the side chains from both N130 and D133 are only involved in intramolecular hydrogen bonds. It has previously been shown that a PvRON2 peptide binds well to PfAMA1 [22]. With this in mind, it is noted that the region corresponding to N130–D133 in PvAMA1 is not conserved in PfAMA1, and that this region is also not involved in the binding between PvRON2 and PfAMA1 (PDB structure 5NQF) [22]. These observations, coupled with the observation that a number of polymorphisms fall within this region, suggests that these residues are not a major determinant for binding to PvRON2 and are hence amenable to polymorphic variation.
There were 7 polymorphic residues observed within the PvDBP dimerization and DARC binding interface, although again most of these were present at low frequencies. The highest frequency polymorphism within this region is R263S, with a minor allele frequency of 20%. In the PDB structure 4NUU, the backbone of R263 is involved in hydrogen bonds to DARC, while the side chain is involved in a single intra-molecular hydrogen bond. This residue is also part of a loop which is disordered when not bound to DARC (loop 254–267) [39], suggesting a level of structural plasticity. The other polymorphic residue of note is T359R, which was identified as a major contact within the DARC binding interface by Batchelor et al. [39], although this polymorphism was observed at a relatively low minor allele frequency of 4.1% across all populations. However, this residue is not conserved between PvDBP and PkDBPα; PkDBPα has an arginine at this position and is also able to bind DARC, supporting the viability of the T359R mutation with regards to maintaining DARC binding ability.
Comparison between 3D and linear sliding window approaches
While a number of previous studies have utilized a linear sliding window approach when calculating Tajima’s D or nucleotide diversity [4,5,6,7, 65], this study is one of the first to utilize a spatially derived Tajima’s D. As such, the Tajima’s D values obtained using a 3D sliding window were compared with those obtained using a conventional linear sliding window, using a window size of 102 base pairs and a step size of 3 base pairs for the linear sliding window (Additional files 9, 10). In general there was broad correspondence between the two approaches for both PvDBP RII and PvAMA1, although there were some additional regions within PvAMA1 with Tajima’s D values above the threshold for significance when using a 3D sliding window approach that were not identified with a linear sliding window. However, these regions were generally structurally contiguous with other stretches of sequence that were above the threshold for significance when using a linear sliding window.
Comparison of spatially derived nucleotide diversity and Tajima’s D between populations
When examining differences in the patterns of nucleotide diversity and Tajima’s D values between various populations worldwide, it appeared that most populations had similar structural patterns of diversity/selection, with a few exceptions that could be due to other population effects such as recent bottleneck and expansion events. To quantify the degree of similarity between structural patterns of selection, we computed Spearman’s rank correlation coefficient for nucleotide diversity and Tajima’s D values between every pair of populations (Fig. 6). Structural patterns of nucleotide diversity were generally highly correlated between populations, with the exception of South Korea for PvAMA1, and Myanmar and Papua New Guinea for PvDBP. In contrast, there was less agreement in the structural patterns of Tajima’s D over each structure. There was reasonable positive correlation between most populations for spatially derived Tajima’s D over the PvAMA1 structure, with the exception of South Korean and Sri Lankan populations. For PvDBP, the Papua New Guinean population was the major outlier when considering spatially derived Tajima’s D values, with the highest values observed on subdomain 3, furthest from the dimerization interface. The lower level of apparent immune selection pressure on PvDBP may explain the more discordant results observed between populations for PvDBP; nearly all Tajima’s D values observed for PvDBP do not meet the threshold for statistical significance (p < 0.05) as defined by Tajima [60] and, therefore, tests of correlation are more sensitive to small variations in Tajima’s D. In contrast, a number of populations had significantly positive spatially derived Tajima’s D values for PvAMA1, including both Papua New Guinean populations (Madang, East Sepik) and the Venezuelan population (Additional file 7).
Similarity in structural patterns of nucleotide diversity and Tajima’s D between populations. Spearman’s rank correlation coefficient was calculated for each pair of populations, comparing spatially derived nucleotide diversity and Tajima’s D values between each residue in the respective protein structures
Discussion
In this study, patterns of nucleotide diversity and selection were examined over the protein structures for the P. vivax antigens PvAMA1 and PvDBP. A number of major observations stand out from this work. Firstly, patterns of diversity on both PvAMA1 and PvDBP were remarkably similar across multiple geographic populations, despite phylogenetic trees for both PvAMA1 and PvDBP sequences suggesting a level of clustering according to geographic location. The only exception to this for PvAMA1 was the South Korean population which displayed evidence of a recent bottleneck and expansion. This similarity in patterns of diversity is important when trying to extend conclusions made from studies from single geographic locations to a worldwide population, and these observations suggest a universality with regards to major epitopes on these antigens. It is also interesting to note that highly polymorphic residues for both PvAMA1 and PvDBP tended to fall within regions surrounding, but not a part of, ligand binding interfaces. For AMA1, RON2 binds in a hydrophobic cleft in DI, and polymorphic residues fall on one side of this hydrophobic cleft, but generally not within residues that make contact with the RON2 peptide. Similarly with PvDBP, contact with DARC occurs primarily via subdomain 2, and the most polymorphic regions were near the DARC binding and dimerization interface. In contrast, the residues directly involved in the PvDBP binding interface were highly conserved. These results are suggestive of two things. Firstly, the residues that make up the key binding interfaces in these two antigens have limited capacity for polymorphic variation due to functional constraints, as has been previously suggested by other studies [39, 63]. This makes them attractive vaccine targets, as potential epitopes within these binding sites would have very limited antigenic diversity, and are also less likely to undergo extensive mutations to evade immune responses. Secondly, the high degree of polymorphism around these interfaces suggests that antibody responses that target these polymorphic sites are capable of inhibiting parasite invasion. This inhibition is likely the result of steric hindrance preventing receptor binding and/or dimerization. Future efforts could involve epitope focusing techniques [66, 67] to direct antibody responses to these key conserved interfaces.
For PvAMA1, we observed balancing selection primarily on DI in all populations examined. Additionally, while the 3D sliding window approach highlighted additional residues under balancing selection as compared to a linear sliding window approach, nearly all of these regions fell within DI. This agrees with a number of other studies in which PvAMA DI is the only domain found to be under significant balancing selection [4, 8,9,10]. This is in contrast to selection pressures observed on PfAMA1, in which both DI and DIII have been observed to be under balancing selection [5,6,7,8,9, 68]. Previous work of ours has applied spatially-derived Tajima’s D calculations over a PfAMA1 structure and identified strong balancing selection in a region bordering DII and DIII [12], lending further evidence to DIII being under immune selection pressure in P. falciparum but not P. vivax AMA1. The biological reasons for such a difference are unclear, as AMA1 has a conserved role between Plasmodium species, although it is possible that DIII of PvAMA1 is less immunogenic than the corresponding PfAMA1 domain due to structural or sequence differences between the two antigens.
Although individuals in malaria endemic areas develop antibodies to PvAMA1 [69,70,71], there are no comprehensive studies on how these antibodies interact with the different domains of PvAMA1. However, AMA1 is functionally conserved across Plasmodium species, and there is evidence that PvAMA1 is functionally equivalent in a P. falciparum transgenic line in which PfAMA1 is replaced by PvAMA1 [72]. As such, comparisons can be drawn with antibody studies on PfAMA1. Dutta et al. [24] generated a panel of monoclonal antibodies (mAbs) to PfAMA1 and observed that their strain specificity and functional activity was determined by the diversity of the epitope sequence. The limited diversity in PvAMA1 DIII observed in this current study aligns with the observations that mAbs to PfAMA1 DIII were the most strain transcending. Similarly, we observed that the polymorphic face of PvAMA1 DI had the highest diversity, in line with the observation by Dutta et al. that mAbs that bound the polymorphic face of PfAMA1 DI were strain specific, compared to the others that bound the conserved face [24]. Importantly, mAbs that bound to the conserved face of PfAMA1 still showed strong growth inhibitory activity suggesting that epitopes on the conserved face can be targets of neutralizing antibodies, despite being under less immune pressure than immunodominant polymorphic regions.
For PvDBP, one of the regions of high diversity across all populations was a previously identified epitope within subdomain 2 termed the DEK epitope [33]. Others have observed that the DEK epitope is highly polymorphic and immunodominant [44]. However, due to the polymorphic nature of this epitope, cross-strain specificity is a concern when creating a PvDBP RII-based vaccine. Recent work has characterized the location of several conserved epitopes within PvDBP RII that are the target of inhibitory mAbs 2D10, 2H2 and 2C6 [73, 74], and all of these conserved epitopes fall within subdomain III, which had the lowest overall nucleotide diversity in this current study. This highlights some of the limitations of using population-level genomic data for identification of functionally important targets of antibody responses—the possibility that conserved regions may contain potential epitopes that are the targets of inhibitory antibodies cannot be excluded.
Several attempts have been made to divert immune responses away from these highly polymorphic regions of PvDBP RII and towards conserved epitopes. One attempt to focus away from polymorphic regions involved mutating residues in the DEK epitope to reduce its immunogenicity, and these DEKnull mutants induce antibodies that bind PvDBP and can inhibit the interaction with Duffy Binding Ligand [33]. More recently, further epitope focusing techniques have been employed with PvDBP RII, with one strategy involving mutation of all polymorphic residues to alanine, threonine or serine residues [75]. This ‘DEKnull-2’ recombinant PvDBP RII construct was shown to elicit broadly neutralizing antibodies following mouse immunization, with some naturally exposed individuals also shown to recognize the conserved epitopes on this construct [75]. Other recent efforts towards the development of a PvDBP RII vaccine include a Phase 1a trial of a prime-boost viral-vectored vaccine that demonstrated both safety and immunogenicity, with cross-strain inhibition demonstrated for the single heterologous strain tested [76].
Given the success using epitope focusing techniques for PvDBP RII, we suggest that such an approach could be applied to AMA1 to focus antibody responses towards conserved epitopes on the silent face of DI or within DIII. This would involve mutating major polymorphic residues to reduce the immunodominance of epitopes within the polymorphic face of DI; the most polymorphic residues identified in this study (Additional file 11) could serve as starting point for this work. Alternatively, it has been shown for PfAMA1 that immunization with multiple heterologous strains of PfAMA1 is capable of inducing strain transcending antibody responses [24], and this approach could also be applied to PvAMA1. Other epitope-focusing approaches also exist, including the use of small protein scaffolds to mimic native epitopes [66, 67], although these might be challenging given the discontinuous nature of many potential epitopes within AMA1. The approaches used in this work could also be applied to other antigens such as PvRBP2b, which has recently been identified as a ligand for reticulocyte invasion via binding to transferrin receptor 1 (TfR1) [77].
Conclusions
In this work, signatures of diversity and selection were identified on PvAMA1 and PvDBP, and it was shown that the regions of high diversity and balancing selection on the protein structure are remarkably similar across a number of populations. This suggests that dominant epitopes are the same across multiple human populations, which has positive implications for the development of a universal P. vivax vaccine. Furthermore, polymorphisms were observed to cluster around binding interfaces on both PvDBP and PvAMA1, suggesting a level of immune pressure on residues surrounding these key binding interfaces. Large regions with very low diversity were also identified for both antigens, and it is suggested that these areas may also be useful targets to focus on for further vaccine development, given previous evidence of functional antibody responses against these conserved regions.
References
WHO. World Malaria Report 2017. Geneva: World Health Organization; 2017. http://www.who.int/malaria/publications/world-malaria-report-2017/en/ Accessed 21 Dec 2017.
Mobegi VA, Duffy CW, Amambua-Ngwa A, Loua KM, Laman E, Nwakanma DC, et al. Genome-wide analysis of selection on the malaria parasite Plasmodium falciparum in West African populations of differing infection endemicity. Mol Biol Evol. 2014;31:1490–9.
Amambua-Ngwa A, Tetteh KKA, Manske M, Gomez-Escobar N, Stewart LB, Elizabeth Deerhake M, et al. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 2012;8:e1002992.
Arnott A, Mueller I, Ramsland PA, Siba PM, Reeder JC, Barry AE. Global population structure of the genes encoding the malaria vaccine candidate, Plasmodium vivax apical membrane antigen 1 (Pv AMA1). PLoS Negl Trop Dis. 2013;7:e2506.
Osier FHA, Weedall GD, Verra F, Murungi L, Tetteh KKA, Bull P, et al. Allelic diversity and naturally acquired allele-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 in Kenya. Infect Immun. 2010;78:4625–33.
Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165:555–61.
Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics. 2001;158:1505–12.
Arnott A, Wapling J, Mueller I, Ramsland PA, Siba PM, Reeder JC, et al. Distinct patterns of diversity, population structure and evolution in the AMA1 genes of sympatric Plasmodium falciparum and Plasmodium vivax populations of Papua New Guinea from an area of similarly high transmission. Malar J. 2014;13:233.
Ord RL, Tami A, Sutherland CJ. Ama1 genes of sympatric Plasmodium vivax and P. falciparum from Venezuela differ significantly in genetic diversity and recombination frequency. PLoS ONE. 2008;3:e3366.
Zakeri S, Sadeghi H, Mehrizi AA, Djadid ND. Population genetic structure and polymorphism analysis of gene encoding apical membrane antigen-1 (AMA-1) of Iranian Plasmodium vivax wild isolates. Acta Trop. 2013;126:269–79.
Premaratne PH, Aravinda BR, Escalante AA, Udagama PV. Genetic diversity of Plasmodium vivax Duffy Binding Protein II (PvDBPII) under unstable transmission and low intensity malaria in Sri Lanka. Infect Genet Evol. 2011;11:1327–39.
Guy AJ, Irani V, Beeson JG, Webb B, Sali A, Richards JS, et al. Proteome-wide mapping of immune features onto Plasmodium protein three-dimensional structures. Sci Rep. 2018;8:4355.
Peterson MG, Marshall VM, Smythe JA, Crewther PE, Lew A, Silva A, et al. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol Cell Biol. 1989;9:3151–4.
Healer J, Crawford S, Ralph S, McFadden G, Cowman AF. Independent translocation of two micronemal proteins in developing Plasmodium falciparum merozoites. Infect Immun. 2002;70:5751–8.
Lamarque M, Besteiro S, Papoin J, Roques M, Normand BV-L, Morlon-Guyot J, et al. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog. 2011;7:e1001276.
Richard D, MacRaild CA, Riglar DT, Chan J-A, Foley M, Baum J, et al. Interaction between Plasmodium falciparum apical membrane antigen 1 and the rhoptry neck protein complex defines a key step in the erythrocyte invasion process of malaria parasites. J Biol Chem. 2010;285:14815–22.
Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, Moch JK, et al. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proc Natl Acad Sci USA. 2011;108:13275–80.
Mugyenyi CK, Elliott SR, McCallum FJ, Anders RF, Marsh K, Beeson JG. Antibodies to polymorphic invasion-inhibitory and non-inhibitory epitopes of Plasmodium falciparum apical membrane antigen 1 in human malaria. PLoS ONE. 2013;8:e68304.
Hodder AN, Crewther PE, Anders RF. Specificity of the protective antibody response to apical membrane antigen 1. Infect Immun. 2001;69:3286–94.
Fowkes FJI, Richards JS, Simpson JA, Beeson JG. The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: a systematic review and meta-analysis. PLoS Med. 2010;7:e1000218.
Pizarro JC, Vulliez-Le Normand B, Chesne-Seck M-L, Collins CR, Withers-Martinez C, Hackett F, et al. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science. 2005;308:408–11.
Vulliez-Le Normand B, Saul FA, Hoos S, Faber BW, Bentley GA. Cross-reactivity between apical membrane antigen 1 and rhoptry neck protein 2 in P. vivax and P. falciparum: a structural and binding study. PLoS ONE. 2017;12:e0183198.
Dutta S, Dlugosz LS, Clayton JW, Pool CD, Haynes JD, Gasser RA 3rd, et al. Alanine mutagenesis of the primary antigenic escape residue cluster, c1, of apical membrane antigen 1. Infect Immun. 2010;78:661–71.
Dutta S, Dlugosz LS, Drew DR, Ge X, Ge X, Ababacar D, et al. Overcoming antigenic diversity by enhancing the immunogenicity of conserved epitopes on the malaria vaccine candidate apical membrane antigen-1. PLoS Pathog. 2013;9:e1003840.
Chenet SM, Tapia LL, Escalante AA, Durand S, Lucas C, Bacon DJ. Genetic diversity and population structure of genes encoding vaccine candidate antigens of Plasmodium vivax. Malar J. 2012;11:68.
Kang J-M, Lee J, Cho P-Y, Moon S-U, Ju H-L, Ahn SK, et al. Population genetic structure and natural selection of apical membrane antigen-1 in Plasmodium vivax Korean isolates. Malar J. 2015;14:455.
Moon S-U, Na B-K, Kang J-M, Kim J-Y, Cho S-H, Park Y-K, et al. Genetic polymorphism and effect of natural selection at domain I of apical membrane antigen-1 (AMA-1) in Plasmodium vivax isolates from Myanmar. Acta Trop. 2010;114:71–5.
Wertheimer SP, Barnwell JW. Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Exp Parasitol. 1989;69:340–50.
Horuk R, Chitnis CE, Darbonne WC, Colby TJ, Rybicki A, Hadley TJ, et al. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261:1182–4.
Cavasini CE, de Mattos LC, Couto AAD, Bonini-Domingos CR, Valencia SH, de Neiras WC, et al. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans R Soc Trop Med Hyg. 2007;101:1042–4.
Ménard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA. 2010;107:5967–71.
Ntumngia FB, Thomson-Luque R, de Torres LM, Gunalan K, Carvalho LH, Adams JH. A novel erythrocyte binding protein of Plasmodium vivax suggests an alternate invasion pathway into Duffy-positive reticulocytes. MBio. 2016;7:e011261.
Ntumngia FB, Adams JH. Design and immunogenicity of a novel synthetic antigen based on the ligand domain of the Plasmodium vivax Duffy binding protein. Clin Vaccine Immunol. 2012;19:30–6.
Adams JH, Blair PL, Kaneko O, Peterson DS. An expanding EBL family of Plasmodium falciparum. Trends Parasitol. 2001;17:297–9.
Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–63.
Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–4.
Maier AG, Duraisingh MT, Reeder JC, Patel SS, Kazura JW, Zimmerman PA, et al. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med. 2003;9:87–92.
Mayer DCG, Cofie J, Jiang L, Hartl DL, Tracy E, Kabat J, et al. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci USA. 2009;106:5348–52.
Batchelor JD, Malpede BM, Omattage NS, DeKoster GT, Henzler-Wildman KA, Tolia NH. Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS Pathog. 2014;10:e1003869.
Batchelor JD, Zahm JA, Tolia NH. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol. 2011;18:908–14.
Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–4.
Cole-Tobian JL, Michon P, Biasor M, Richards JS, Beeson JG, Mueller I, et al. Strain-specific Duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous Plasmodium vivax strains in Papua New Guinean children. Infect Immun. 2009;77:4009–17.
Xainli J, Baisor M, Kastens W, Bockarie M, Adams JH, King CL. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J Immunol. 2002;169:3200–7.
Chootong P, Ntumngia FB, VanBuskirk KM, Xainli J, Cole-Tobian JL, Campbell CO, et al. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect Immun. 2010;78:1089–95.
Grimberg BT, Udomsangpetch R, Xainli J, McHenry A, Panichakul T, Sattabongkot J, et al. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 2007;4:e337.
Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–43.
Sukumaran J, Holder MT. DendroPy: a Python library for phylogenetic computing. Bioinformatics. 2010;26:1569–71.
Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Le SV. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22.
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9.
Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5.
Eswar N, John B, Mirkovic N, Fiser A, Ilyin VA, Pieper U, et al. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31:3375–80.
Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815.
Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–74.
Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9:90–5.
van der Walt S, Colbert SC, Varoquaux G. The NumPy array: a structure for efficient numerical computation. Comput Sci Eng. 2011;13:22–30.
Schrödinger LLC. The PyMOL molecular graphics system, version 1.8. 2015.
Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95.
Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709.
Faber BW, Kadir KA, Rodriguez-Garcia R, Remarque EJ, Saul FA, Normand BV-L, et al. Low levels of polymorphisms and no evidence for diversifying selection on the Plasmodium knowlesi apical membrane antigen 1 Gene. PLoS ONE. 2015;10:e0124400.
Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, et al. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci USA. 2005;102:12736–41.
Takala SL, Coulibaly D, Thera MA, Batchelor AH, Cummings MP, Escalante AA, et al. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci Transl Med. 2009;1:2ra5.
Escalante AA, Grebert HM, Chaiyaroj SC, Magris M, Biswas S, Nahlen BL, et al. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol Biochem Parasitol. 2001;113:279–87.
Oscherwitz J. The promise and challenge of epitope-focused vaccines. Hum Vaccin Immunother. 2016;12:2113–6.
Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507:201.
Mehrizi AA, Sepehri M, Karimi F, Djadid ND, Zakeri S. Population genetics, sequence diversity and selection in the gene encoding the Plasmodium falciparum apical membrane antigen 1 in clinical isolates from the south-east of Iran. Infect Genet Evol. 2013;17:51–61.
Fowkes FJI, McGready R, Cross NJ, Hommel M, Simpson JA, Elliott SR, et al. New insights into acquisition, boosting, and longevity of immunity to malaria in pregnant women. J Infect Dis. 2012;206:1612–21.
Cutts JC, Powell R, Agius PA, Beeson JG, Simpson JA, Fowkes FJI. Immunological markers of Plasmodium vivax exposure and immunity: a systematic review and meta-analysis. BMC Med. 2014;12:150.
McLean ARD, Boel M, McGready R, Ataide R, Drew D, Tsuboi T, et al. Antibody responses to Plasmodium falciparum and Plasmodium vivax and prospective risk of Plasmodium spp. infection postpartum. Am J Trop Med Hyg. 2017;96:1197–204.
Drew DR, Sanders PR, Weiss G, Gilson PR, Crabb BS, Beeson JG. Functional conservation of the AMA1 host-cell invasion ligand between P. falciparum and P. vivax: a novel platform to accelerate vaccine and drug development. J Infect Dis. 2018;217:498–507.
Chen E, Salinas ND, Huang Y, Ntumngia F, Plasencia MD, Gross ML, et al. Broadly neutralizing epitopes in the Plasmodium vivax vaccine candidate Duffy Binding Protein. Proc Natl Acad Sci USA. 2016;113:6277–82.
Ntumngia FB, Schloegel J, Barnes SJ, McHenry AM, Singh S, King CL, et al. Conserved and variant epitopes of Plasmodium vivax Duffy binding protein as targets of inhibitory monoclonal antibodies. Infect Immun. 2012;80:1203–8.
Ntumngia FB, Pires CV, Barnes SJ, George MT, Thomson-Luque R, Kano FS, et al. An engineered vaccine of the Plasmodium vivax Duffy binding protein enhances induction of broadly neutralizing antibodies. Sci Rep. 2017;7:13779.
Payne RO, Silk SE, Elias SC, Milne KH, Rawlinson TA, Llewellyn D, et al. Human vaccination against Plasmodium vivax Duffy-binding protein induces strain-transcending antibodies. JCI Insight. 2017;2:93683 (Epub ahead of print).
Gruszczyk J, Kanjee U, Chan L-J, Menant S, Malleret B, Lim NTY, et al. Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science. 2018;359:48–55.
Authors’ contributions
AJG designed the study, performed the research, analysed the results and wrote the manuscript; PAR contributed the modelled PvAMA1 structure; VI, JSR and PAR provided critical interpretation of the data. All authors revised, commented and read the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
Funding was provided by the National Health and Medical Research Council (NHMRC) of Australia including an Early Career Research Fellowship (APP1037722) and Project Grant (APP1125788) to JSR; and an Australian Postgraduate Award to support AJG through Monash University. Burnet Institute received funding from the NHMRC Independent Research Institutes Infrastructure Support Scheme, and the Victorian State Government Operational Infrastructure Support Scheme.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1.
Domains/subdomains of PvAMA1.
Additional file 2.
Domains/subdomains of PvDBP RII.
Additional file 3.
Spatially-derived nucleotide diversity for PvAMA1 across multiple populations.
Additional file 4.
Spatially-derived Tajima’s D for PvAMA1 across multiple populations.
Additional file 5.
Spatially-derived nucleotide diversity for PvDBP across multiple populations.
Additional file 6.
Spatially-derived Tajima’s D for PvDBP across multiple populations.
Additional file 7.
Location of statistically significant (p < 0.05) Tajima’s D values on modelled PvAMA1 (a) and PvDBP (b) structures.
Additional file 8.
Sequence diversity at interfaces.
Additional file 9.
Comparison of spatially derived Tajima’s D and conventional linear sliding window calculation of Tajima’s D for PvAMA1.
Additional file 10.
Comparison of spatially derived Tajima’s D and conventional linear sliding window calculation of Tajima’s D for PvDBP RII.
Additional file 11.
Table of highly polymorphic residues within PvAMA1.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Guy, A.J., Irani, V., Richards, J.S. et al. Structural patterns of selection and diversity for Plasmodium vivax antigens DBP and AMA1. Malar J 17, 183 (2018). https://doi.org/10.1186/s12936-018-2324-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-018-2324-3