Abstract
Background
Identifying Plasmodium vivax antigen-specific antibodies associated with P. vivax infection and protective immunity is key to the development of serosurveillance tools and vaccines for malaria. Antibody targets of P. vivax can be identified by seroepidemiological studies of individuals living in P. vivax-endemic areas, and is an important strategy given the limited ability to culture P. vivax in vitro. There have been numerous studies investigating the association between P. vivax antibody responses and P. vivax infection, but there has been no standardization of results to enable comparisons across populations.
Methods
We performed a systematic review with meta-analysis of population-based, cross-sectional, case-control, and cohort studies of individuals living in P. vivax-endemic areas. We searched 6 databases and identified 18 studies that met predefined inclusion and quality criteria, and examined the association between antibody responses to P. vivax antigens and P. vivax malaria.
Results
The majority of studies were published in South America (all from Brazil) and the rest from geographically diverse areas in the Asia-Pacific region. Considerable heterogeneity in estimates was observed, but IgG responses to PvCSP, PvMSP-119, PvMSP-9RIRII, and PvAMA1 were associated with increased odds of P. vivax infection in geographically diverse populations. Potential sources of heterogeneity included study design, different transmission intensities and transmigrant populations. Protective associations were observed for antibodies to PvMSP-119, PvMSP-1NT, PvMSP-3α and PvMSP-9NT antigens, but only in single geographical locations.
Conclusions
This systematic review revealed several antigen-specific antibodies that were associated with active infection and protective immunity, which may be useful biomarkers. However, more studies are needed on additional antigens, particularly cohort studies to increase the body of evidence for protective immunity. More studies representing diverse geographical regions encompassing varying P. vivax endemicities are needed to validate the generalizability of the findings and to provide a solid evidence base for the use of P. vivax antigens in vaccines and serosurveillance tools.
Similar content being viewed by others
Background
Plasmodium vivax is the most widely distributed species of human malaria, with an estimated 40% of the world's population being at risk of P. vivax infection [1]. The majority of P. vivax infections occur in Central and South-East Asia, and there are approximately 80 to 300 million clinical cases of P. vivax malaria each year [1]. Despite the large burden of disease, P. vivax has traditionally been neglected because it has been considered a relatively benign form of malaria. Now it is recognized that P. vivax can cause severe disease (similar to that of Plasmodium falciparum) [2] and, together with increased recognition of the burden, there are renewed efforts in the development of P. vivax-specific interventions (that is, vaccines) and surveillance tools (diagnostics, serosurveillance) to expedite the goal of malaria elimination and eradication [3],[4].
Currently, only two P. vivax vaccine candidates (Pv Duffy binding protein (PvDBP) and Pv circumsporozoite protein PvCSP) are in clinical trials (Phase I) compared with 23 P. falciparum vaccine candidates (including one in Phase III trials: RTS,S) [5],[6]. This may reflect the previous neglect of P. vivax, the difficulty in maintaining P. vivax in culture, and the limited animal models of infection currently available. Such technical challenges have hindered the ability to prioritize P. vivax candidate antigens against pre-clinical selection criteria, including knowledge of protein function and antigenic diversity, and demonstrations that antibodies against an antigen inhibit growth in vitro or function in other ways, or are protective in animal models of infection [7]. In the absence of an in vitro system, P. vivax antigens can be selected based on P. falciparum homologues and an additional pre-clinical selection criterion, namely, that the antigen induces naturally acquired immunity in individuals living in malaria-endemic areas [7].
In P. vivax-endemic areas, the prevalence and density of P. vivax infection and the incidence of P. vivax symptomatic malaria decrease with age [8]. This epidemiological pattern reflects the acquisition of natural immunity that develops after repeated exposure [9]. This immunity is non-sterilizing and does not protect against infection, but acts by reducing parasite numbers in the blood and the subsequent clinical symptoms. Antibodies are thought to be an important component of naturally acquired immunity, and are considered to be biomarkers of both immunity and exposure. Potential antibody targets include P. vivax antigens expressed on sporozoites (the pre-erythrocytic liver stage), the invading merozoite and the surface of infected erythrocytes (erythrocytic stage) and the gametocyte (sexual stage) [8]. P. vivax also has an additional dormant stage in the liver, the hypnozoite, which is believed to be responsible for relapses in P. vivax infection [10].
There have been numerous studies investigating associations between P. vivax immune responses and P. vivax infection, but there is considerable heterogeneity between studies, both in terms of methodology and presentation of results, making cross-study comparison problematic. Here, we aimed to review and synthesize the literature, by standardizing analyses and identifying targets of naturally acquired immunity to P. vivax, which we have previously done similarly for P. falciparum[11]. There were two key objectives of this study: to determine antigen-specific antibody responses associated with infection, and to determine antibody responses associated with protective immunity. We included cross-sectional and case-control studies in order to identify markers of P. vivax infection, and also included cohort studies, which provide the highest level of evidence to detect causal effect in observational research, in order to identify antibody responses that protect against P. vivax malaria. The overarching aim of the study was to provide a more comprehensive understanding of antibody-mediated immunity to P. vivax and, more specifically, to help inform the development of vaccines and serosurveillance tools to facilitate the control, elimination and eradication of P. vivax.
Methods
The Meta-analysis of Observational Studies in Epidemiology (MOOSE) working group [12] guidelines and the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) specifications were adhered to in the conducting and reporting of this systematic review and meta-analysis [13]. For a completed PRISMA checklist, see Additional file 1.
Search methods for identification of studies
PubMed, Web of Science, Scopus, Google Scholar, African Index Medicus, and the Latin American and Caribbean Health Sciences Literature (LILACS) databases were searched for studies published in all years up to and including 30 November 2013 that examined the association of antibody responses to P. vivax antigens with P. vivax infection or P. vivax malaria. Key words included: malaria, vivax, plasmodium, immunoglobulin, IgG, antibody, immunity, rhoptry, microneme, sporozoite, CSP, circumsporozoite, TRAP, thrombospondin, merozoite, MSP, AMA, DBP, Duffy binding protein, EBA, EBP, erythrocyte binding*, EMP, erythrocyte membrane protein, RBL, reticulocyte binding like protein, RBP, reticulocyte binding protein, VSA, variant surface antigen, VIR, gameotocyte, transmission blocking, Pvs25, ookinete surface protein, Pvs28, sexual stage surface protein, transmission-blocking target antigen, and Pvs230. The full search strategy for one database (PubMed) is provided (see Additional file 2). The reference lists of the obtained papers were searched for further studies. Studies reported in languages other than English were included, and were translated into English using online translation applications. A priori, we did not formally attempt to identify unpublished population studies because this would have required us to provide substantial descriptions of the study design, sample testing, and analysis used in the studies, and a review of ethical and other issues.
Criteria for considering studies
Study designs and study participants
Population-based cross-sectional, case-control, and cohort studies, including treatment to re-infection studies, were included in the systematic review. Randomized controlled trials and vaccine efficacy trials of blood-stage vaccines were excluded because rigorous inclusion and exclusion criteria are applied in these studies, thus the participants are, typically, not representative of the general population. The primary criterion for study inclusion was inclusion of individuals (children, adults, and/or pregnant women) living in P. vivax-endemic areas. All geographical locations were included. Studies of the following types of populations were excluded because they were unlikely to represent the general population: populations experiencing epidemic malaria (that is, no previous exposure to P. vivax); returned travellers; military personnel; and populations in which greater than 20% of individuals were transmigrants who had resided in the area for less than 5 years at the time of sampling. Studies that included multiple population subsets were assessed on a sub-population basis to determine eligibility for inclusion.
Antibody measures
Studies that measured total immunoglobulin G (IgG), total IgM, or IgG subclass (1-4) responses to recombinant or synthetic defined P. vivax antigens were considered. Antibody responses to full-length proteins, processing products, and defined regions of P. vivax antigens from any life-cycle stage and any subcellular location were also included. Responses to peptides representing undefined regions or incomplete domains or subdomains of antigens were excluded, with the exception of proteins or defined domains that could not be expressed as a single product. In such cases, responses to a combination of protein fragments representing the full-length protein or domain were analyzed. For cohort studies, if antibody responses were measured at multiple time points the baseline (that is, time 0), antibody responses were analyzed. Data from cohort studies in which antibody responses were determined after malariometric measures were excluded.
Malaria outcome measures
The following malaria outcome measures were included: P. vivax infection, high-density P. vivax infection, and symptomatic P. vivax malaria, using the definitions as described in the individual studies. In cohort studies, P. vivax re-infection was also included as an outcome. Studies in which malariometric measures were determined retrospectively (for example, where cumulative history of malaria exposure was the exposure or outcome of interest) were excluded.
Quality criteria
The minimum quality criteria for inclusion were: confirmation of P. vivax parasitemia by light microscopy, rapid detection kit, or PCR; detection of P. vivax malaria by active and/or passive case detection; and symptomatic malaria defined by fever and/or history of fever (within the past 72 hours) plus P. vivax parasitemia. In studies in which symptomatic malaria was the outcome of interest, cases of symptomatic malaria in individuals with P. falciparum and P. vivax co-infection were excluded because the symptoms could not be attributed exclusively to one or the other species. Cut-offs for positive antibody responses by ELISA had to be defined by the use of unexposed (malaria-naïve) controls rather than individuals from the same exposed population found to be P. vivax-negative at the time of sampling. For treatment to re-infection studies, if treatment failure was accounted for, it had to be defined by either genetic analysis or documented clearance of infection within a specified time frame appropriate for the chosen antimalarial. In case-control studies, at least one control for every case had to be recruited from the same population (that is, studies that recruited a small number of laboratory controls for antibody comparison purposes were excluded).
Selection of studies
Two independent review authors used the inclusion and exclusion criteria to screen titles and abstracts. The full text of potentially relevant studies was retrieved and examined for compliance with the inclusion and exclusion criteria by the same two review authors independently. Discrepancies were resolved by discussion with a third author.
Effort to include all available studies and data
Authors of original studies were contacted if relevant information on the study population, eligibility criteria, or key study data were not presented in the published report. For studies in which antibody responses to P. vivax antigens were described, but no details of P. vivax outcomes were reported, authors were invited to provide malariometric data to enable the study to be included in the review. If authors were unable to provide estimates or data, the study was classified as not meeting the inclusion and/or quality criteria, and was excluded from the systematic review. Cross-sectional data from cohort studies was extracted for inclusion in cross-sectional analyses. In studies in which multiple cross-sectional surveys were performed in the same population, estimates were reported for individual surveys if the data were available. For studies in which antibody responses were analysed as the outcome variable, data were re-analyzed so that malaria or P. vivax infection was the outcome variable.
Risk of bias in individual studies
At an individual study level, selection bias was assessed to determine whether participants were representative of the general population by reviewing individual study inclusion and exclusion criteria. Selection bias in case-control studies was assessed by assessing the comparability of cases and controls as part of the systematic review quality criteria. We excluded case-control studies if the source population differed between cases and controls, because bias would be introduced into the estimates of the association between P. vivax antibodies and outcomes. An additional selection bias can occur in case-control studies when cases and/or controls are selected based on criteria relating to their exposure (that is, antibody) status or when there are differences in the reporting of exposure between cases and controls. However, this is unlikely because immunoassays would be performed after enrolment into the study. Information bias (resulting from flaws in measuring antibody and P. vivax outcome data) are unlikely because antibodies are measured using immunoassays that are standardized within each study and across outcome groups. The quality criterion of this review ensured accurate measurement of P. vivax outcomes, and it is unlikely that measurement of outcomes would differ depending on antibody groups. To reduce bias further, we excluded studies that measured the P. vivax outcome prior to antibody determination. Although these studies may be useful in determining markers of exposure, we excluded such studies because unmeasured P. vivax exposure and/or P. vivax antibody decay between measurements may lead to misclassification and bias in estimates of association. For measures of association, estimates adjusted for demographic variables and/or spatial confounders are reported where possible to reduce confounding. Estimates adjusted for other anti-P. vivax antibodies are not reported because antibody responses are typically highly correlated, making it difficult to estimate their individual regression coefficients reliably [11]; in these cases unadjusted estimates are reported.
Data analysis
Data collection
Measures of association (odds ratio (OR), risk ratio (RRs), incidence rate ratio (IRR), or hazard ratios (HR)) and their 95% confidence intervals (CIs) were extracted or derived using data reported in the publications. Data extraction was performed independently by two reviewers using a proforma. Contact with authors was established through an initial email explaining the nature of the systematic review and the information required, together with the proforma. If the corresponding author did not respond within three email attempts, no further action was taken. Where a study did not provide measures of association (or they could not be calculated using the information provided) the study results were used only for qualitative analysis.
Standardization of antibody measures
Measurement of antibody levels by ELISA does not produce a common metric between studies. Individuals can be classified as `responders’ or `non-responders’ relative to a negative control (unexposed sera) within each study. Study-specific comparisons of these exposure variables can then be pooled [11]. However, categories based upon arbitrary cut-offs (including categories of responders based on statistical rankings) cannot be pooled across studies. For studies in which the antibody measures were analyzed as continuous exposure variables, authors were asked to reanalyze their data to provide a binary classification of responders versus non-responders.
Standardization of malaria outcome measures
Studies were grouped according to the study design used to examine the relationship between P. vivax antibodies and P. vivax outcome. For cross-sectional and case-control studies, OR was extracted or calculated, and for cohort studies, RR, HR, and IRR were extracted or calculated where possible, or unadjusted ORs were converted to RR [14] (RR, HR, and IRR are hereinafter denoted as RR). An RR/OR of 1 indicates that the risk/odds of malaria is equal for those with (responders) and those without (non-responders) antibody responses. Separate estimates were obtained for P. vivax detected by light microscopy, PCR, and ligase detection reaction-fluorescent microsphere assay (LDR-FMA). For meta-analyses, estimates using different parasite detection methods were combined. For studies in which multiple estimates were reported for different detection methods, the estimate reflecting the more commonly used method was presented in the forest plot to enable comparisons, and other estimates were presented in the text. Where zero counts were present in 2 × 2 tables, a constant value (0.5) was added to all cells to enable estimation of the OR/RR.
Our aim was to obtain a single estimate for each antibody response and P. vivax outcome. If antibody responses to the same antigen in the same population-based study were reported in several publications, results from the largest sample size were used. Separate estimates were obtained for the OR/RR associated with the PvCSP repetitive domain (VK210, VK247, P. vivax-like alleles, NS1/81-V20 antigen, and VK210 and VK247 chimeric antigen), Pv merozoite surface protein (PvMSP)-119, PvMSP-1 N-terminus, Pv apical membrane antigen (PvAMA1) ectodomain, PvDBP (DBP region II AH, O, P, and Sal 1 alleles, DBP regions II-IV, Sal 1 allele), PvMSP-9 (Block I and Block II repeats, Block II repeats, N-terminal region), PvMSP-3α (full-length, N-terminal region, Block I repeats, Block II repeats), PvMSP-5, P. vivax reticulocyte binding protein (PvRBP1; the extracellular domain was expressed as overlapping recombinant fragments), and PvSERA4. Together with 95% CIs and P values, we interpreted a 20% relative difference in odds/risk of P. vivax outcome to be a clinically meaningful difference between antibody groups a priori, which is approximately half of the observed efficacy of the P. falciparum RTS,S vaccine in Phase III trials [15],[16].
Synthesis of results: meta-analysis
A meta-analysis was performed for each antigen, stratified by study design and P. vivax outcome. Where there were two or more studies that could be combined, a pooled estimate for each outcome was calculated using either a fixed-effects or random-effects model. The standard error of the natural logarithm (ln) of the RR/OR was calculated using the formula: (ln(upper limit of CI) -ln(estimate))/1.96. For fixed-effects models, pooled effects estimates were weighted by the inverse of the individual study standard error. Where random-effects models were specified, a between-study variance component was incorporated into the study weights [17]. Between-study heterogeneity was measured with the I 2 statistic, and represents the percentage of variation in a pooled estimate attributable to between-study variability [18]. Tests for significant between-study heterogeneity were also reported, and were based on the weighted sum of the differences between study estimates and the overall pooled estimate; the statistic takes a χ2 distribution with degrees of freedom equal to the number of studies minus 1 [17]. If heterogeneity was 30% or less, a meta-analysis based on a fixed-effects model was specified; otherwise a random-effects approach was used. Where the heterogeneity exceeded 75% and/or the heterogeneity test statistic was significant at P < 0.1, a pooled effect was not estimated [18]-[21]. Owing to the small number of studies included in the meta-analyses, sensitivity analyses and assessments of publication bias were not performed. All analyses were performed using STATA software (V11; StataCorp, College Station, TX, USA).
Results
Identification and description of included studies
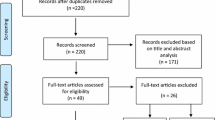
The database searches identified 1,411 records, of which 162 potentially relevant studies were identified, based upon title and abstract. The full texts of these 162 studies were examined to determine whether they complied with eligibility criteria: 114 did not meet the inclusion criteria (see Additional file 3), 7 fulfilled the inclusion and quality criteria (Figure 1), and 41 studies potentially met inclusion and quality criteria. The authors of the 41 studies that potentially met inclusion and quality criteria were contacted, yielding a further 11 studies that met inclusion and quality criteria, providing a total of 18 studies that were included in the review [22]-[39] (Figure 1). Details of these 18 studies are shown in Table 1. Of these 18 studies, 9 were cross-sectional, 6 were cohort (4 of which also provided cross-sectional data), and 3 were case-control studies. One cohort study contributed two publications [34],[35], and one publication provided data from two countries [22]. For the purpose of this review, we shall refer to each publication as a study.
The included studies reported data from Brazil (n = 10), Papua New Guinea (n = 4), Indonesia (n = 2), Thailand (n = 2), and Turkey (n = 1) (Table 1). Sample sizes of the included studies ranged from 61 to 432 study participants, and the majority of studies included both children and adults (4 included children only, 1 included pregnant women only). Antibody responses to P. vivax erythrocytic stage proteins were the main antigens studied (PvMSP-1, n = 8; PvDBP, n = 4; PvMSP-3α, n = 2; PvMSP-5, n = 1; PvMSP-9, n = 2; PvAMA1, n = 2; PvRBP1, n = 1; PvSERA4, n = 1) and three studies examined antibody responses to the pre-erythrocytic stage protein PvCSP (Table 1). No studies examined responses to gametocyte antigens. Details of the recombinant antigens investigated are outlined in detail (see Additional file 4). Total IgG responses were analyzed in 17 studies, with IgM and IgG subclass responses being examined in 4 studies each (Table 1). IgG1 and IgG3 subclasses were the predominant IgG subclass responses in all studies [27],[31],[33],[39],[40] (see Additional file 5). P. vivax infection was the most commonly examined outcome (n = 18), followed by symptomatic P. vivax infection (n = 5) and high-density P. vivax infection (>150 parasites/μl, n = 1). Light microscopy was used for P. vivax detection in the majority of studies (n = 13), with four studies using PCR or microscopy and three using LDR-FMA. For the purpose of the review, all P. vivax infection was diagnosed by light microscopy unless stated otherwise. Results are presented for each antigen stratified by study design: cross-sectional and case-control studies, to identify markers of P. vivax infection, and cohort studies, to identify antibody responses that protect against P. vivax malaria.
Association between antibody responses to PvCSP repeat region and P. vivax
Circumsporozoite protein (CSP), the predominant surface protein of the pre-erythrocytic, sporozoite stage parasite, has been implicated in the invasion of hepatocytes [41]. P. vivax CSP contains a highly immunogenic central repeat domain flanked by amino and carboxyl sequences, which include highly conserved protein stretches (Regions I and II-plus). Three main allelic forms of PvCSP, differing mainly in the central repeat region, have been described: VK210, VK247 [42], and P. vivax-like [43].
Cross-sectional and case-control studies
Two cross-sectional studies [28],[39] and one case-control study [38] examined the association of antibody responses to the PvCSP repeat region and P. vivax infection. A cross-sectional study in Brazil showed no association between IgG responses to the P. vivax-like allele of PvCSP and odds of P. vivax infection (OR = 1.09, responders versus non-responders), but IgG responders to VK210 or VK247 alleles had a non-significant reduction of 74% and 57% respectively, in the odds of P. vivax infection compared with non-responders (Figure 2) [28]. By contrast, a cross-sectional study in Turkey showed a 2.4-fold increase in the odds of P. vivax infection in IgG responders to combined VK210 and VK247 alleles (compared with non-responders, Figure 2) [39]. Similar associations were also seen for IgM (OR = 3.8, 95% CI 2.06 to 7.01) [39]. A case-control study in Thailand also showed a similar 2.4-fold increase in the odds of P. vivax infection in IgG responders to the NS1/81-V20 antigen, which includes the CSP repetitive domain (VK210 allele) compared with non-responders (Figure 2) [38].
Forest plot of the association of Pv CSP IgG responses with Plasmodium vivax infection. Estimates represent the odds of P. vivax infection in IgG responders compared with non-responders. aData supplied by the original authors and estimate calculated by the current authors; bpublished estimate. All estimates are unadjusted. Abbreviations: LM, light microscopy; W, weight.
Association between antibody responses to PvDBP and Plasmodium vivax
PvDBP is a leading vaccine candidate because invasion of erythrocytes is largely dependent upon its interaction with the Duffy blood-group antigen [44]. The conserved N-terminal cysteine-rich rich region II (PvDBPII) constitutes the receptor binding domain of PvDBP [45],[46]. The full-length ectodomain of PvDBP, comprising regions II to VI, is thought to correspond to the soluble form of the protein [47].
PvDBP region II
Cross-sectional studies
The association of antibodies to PvDBP region II and P. vivax infection was examined in one cross-sectional study and one cohort study [30],[34]. The cross-sectional study was conducted at two study sites in Brazil, and pooled OR showed that IgG responders to PvDBPII Sal 1 strain had higher odds of P. vivax infection compared with non-responders (pooled OR using fixed effects (feOR) = 2.82, 95% CI 0.71 to 11.15, I 2 = 0%) (Figure 3) [30] indicating that this antigen may be indicative of exposure in this population.
Forest plot of the association of Pv DBP IgG responses with Plasmodium vivax infection. Estimates represent the estimate of P. vivax infection in IgG responders compared with non-responders, unless stated otherwise. For cross-sectional studies, the estimate is an odds ratio; for cohort studies, it is a risk ratio. 1Colina study site; 2Ribeirinha study site; 3First (baseline) survey; 4Second survey. aEstimate supplied by the original authors following correspondence; bdata supplied by the original authors and estimate calculated by the current authors; cestimate calculated by the current authors from data in the paper; dpublished estimate. All estimates are unadjusted, with the exception of estimates from Cole-Tobian et al. [34], which were adjusted for age. When I 2 was ≤30%, meta-analysis based on a fixed-effects model was conducted. Estimates for distinct alleles of PvDBPII were not combined in meta-analysis. Abbreviations: AU, antigen units; LM, light microscopy; RTQ-PCR, real-time quantitative polymerase chain reaction; PNG, Papua New Guinea; W, weight.
Cohort studies
A cohort study conducted in PNG by Cole-Tobian et al.[34] revealed no evidence for an association between IgG responses to any of the PvDBPII alleles studied (AH, O, P, or Sal 1) and protection against P. vivax infection detected by light microscopy (log(antigen units + 1); HR ranged from 0.96 to 0.98, Figure 3) or LDR-FMA (log(antigen units + 1); HR ranged from 0.92 to 1.02) [34]. However, IgG responders to PvDBPII (O, P, and Sal 1 alleles) had a lower risk of high-density P. vivax infections (>150 parasites/μl) compared with non-responders (HR ranged from 0.74 to 0.85) (Figure 3) [34]. In the same Papua New Guinean cohort, King et al.[35] tested plasma samples for their ability to inhibit binding of PvDBPII to its receptor, Duffy antigen: individuals with high-level (>90%) binding inhibitory activity had a 55% reduction in risk of P. vivax infection detected by light microscopy compared with those with low-level (<50%) binding inhibitory activity (HR = 0.45, 95% CI 0.2 to 0.98) [35], providing further evidence of PvDBPII as a target of protective antibodies.
PvDBP region II-IV
Cross-sectional studies
The association of antibodies to PvDBP region II-IV (PvDBPII-IV) and P. vivax infection was examined in one cross-sectional study [23] and one cohort study that also provided two sets of cross-sectional data [29]. In a cross-sectional study, Kano et al.[23] found no evidence for any association between IgG responses to PvDBPII-IV Sal 1 and P. vivax infection. One study by Souza-Silva et al. provided data from two cross-sectional surveys conducted in the same study site in Brazil [29]. While the first (baseline) survey provided no evidence for an association between IgG responses to PvDBPII-IV Sal 1 and P. vivax infection detected by light microscopy (OR = 1.10, 95% CI 0.43 to 2.82), the second survey showed that the IgG responders had increased odds of P. vivax infection compared with non-responders (OR = 4.14, 95% CI 1.58 to 10.8) (Figure 3) [29]. Owing to significant heterogeneity, these estimates were not combined (I 2 = 67.6%, P = 0.046).
Cohort studies
Cohort data from Souza-Silva et al.[29] found a 22% increased prospective risk of P. vivax infection (detected by light microscopy or PCR) in PvDBPII-IV IgG responders compared with non-responders (RR = 1.22, 95% CI 0.73 to 2.06, Figure 3). These results suggest that P. vivax infection during follow-up induced an anti-PvDBPII-IV response.
Association between antibody responses to PvMSP-1 and P. vivax
MSP-1 is conserved in all Plasmodium species, and is thought to be essential for blood-stage development of the parasite [48]. PvMSP-1 has a polymorphic N-terminus (PvMSP-1NT) and a relatively conserved C-terminus [49]. Studies in P. falciparum have established that post-translational proteolytic processing of PfMSP-1 generates four fragments, including a C-terminal 42 kDa fragment, which is further processed into a 19 kDa fragment that remains on the surface of the merozoite during invasion (PvMSP-119) [50]-[52].
PvMSP-119
A total of eight studies investigated responses to PvMSP-119 and P. vivax outcomes [22],[23],[26],[27],[32],[34],[39],[40].
Cross-sectional studies
Four cross-sectional studies (providing five sets of data) investigated the association between IgG responders to PvMSP-119 and P. vivax infection (Figure 4) [22],[23],[26],[39]. Meta-analysis revealed significant heterogeneity between studies (I 2 = 73.8%, P = 0.004), so a pooled estimate was not reported. Increased odds of P. vivax infection (diagnosed by light microscopy or by LM in combination with PCR) in PvMSP-119 IgG responders compared with non-responders was found in Brazil (increased odds of 81% [23] and 18% [26]), Turkey (447% increased odds [39]) and Papua New Guinea (100% increased odds [22]) (Figure 4). Conversely, data from another Brazilian study [22] showed that IgG responders to PvMSP-119 had 48% decreased odds of P. vivax infection (compared with non-responders) [22]. However, when these authors investigated the outcome of symptomatic P. vivax malaria at sites in Brazil and PNG, they found that IgG responders to PvMSP-119 had 51% increased odds of symptomatic P. vivax (feOR =1.51, 95% CI 0.71 to 3.23, I 2 = 0%; Figure 4) [22]. Overall, these data, taken together support IgG response against PvMSP-119 as a marker of P. vivax infection in geographically diverse populations (Figure 4).
Forest plot of the association of Pv MSP-1 19 IgG responses with Plasmodium vivax outcomes. Estimates represent the estimate of P. vivax infection in IgG responders compared with non-responders unless stated otherwise. For cross-sectional and case-control studies, the estimate is an odds ratio; for cohort studies, it is a risk ratio. 1Meta-analysis of IgG responses to PvMSP-119 and odds of P. vivax infection (estimates from cross-sectional studies) showed a high degree of heterogeneity (I 2 = 73.8%, P = 0.004), so results were not pooled. aData supplied by the original authors, and estimate calculated by the current authors; bestimate calculated by the current authors from data in the paper; cpublished estimate. All estimates are unadjusted, with the exception of estimates from Cole-Tobian et al. [34], which were adjusted for age, and Noguiera et al. [27], which were adjusted for geographical sector. When I 2 was ≤30%, meta-analysis based on a fixed-effects model was conducted. AU, antigen units; LM, light microscopy; PCR, polymerase chain reaction; PNG, Papua New Guinea; RTQ-PCR, real-time quantitative PCR; W, weight.
Two studies looked at IgM responses, as a marker of recent exposure to PvMSP-119. Yildiz Zeyrek et al.[39] showed that IgM responses were associated with higher odds of P. vivax infection and of increased magnitude compared with IgG (responders compared with non-responders, OR = 48.8, 95% CI 16.3 to 146.1). One cross-sectional study in Indonesia, which examined combined IgG and IgM responses (and was therefore not included in the IgG or IgM meta-analyses), showed a 71% reduction in the odds of P. vivax infection in responders versus non-responders (OR = 0.29, 95% CI 0.09 to 0.88) [32].
Cohort studies
IgG against PvMSP-119 as a marker of protective immunity was assessed in two cohort studies [27],[34]. No evidence for an association between PvMSP-119 IgG responses and P. vivax infection was found in Brazil (responders versus non-responders; RR = 1.12, 95% CI 0.56 to 2.24) [27] or Papua New Guinea (log(antigen units + 1); light microscopy: RR = 0.92, 95% CI 0.62 to 1.36 (Figure 4); LDR-FMA: RR = 1.07, 95% CI 0.78 to 1.47) [34]. Although Cole-Tobian et al.[34] found no association with P. vivax infection in Papua New Guinea, they did observe a 31% reduced risk of high-density P. vivax infection (≥150 parasites/μl) (log(antigen units + 1); RR = 0.69, 95% CI 0.34 to 1.38).
PvMSP-1 N-terminus
Cross-sectional studies
The cross-sectional study by Fernandez-Becerra et al.[22], conducted in Brazil and PNG, also investigated responses against the N-terminus of PvMSP-1 (PvMSP-1NT) and found no evidence for an association between IgG response (responders versus non-responders) to PvMSP-1NT and either P. vivax infection or symptomatic P. vivax (feOR = 1.19, 95% CI 0.56 to 2.55, I 2 = 0% and feOR = 0.93, 95% CI 0.42 to 2.04, I 2 = 0%, respectively; Figure 5). By contrast, cross-sectional data from Versiani et al.[31] in Brazil showed that IgG responders to PvMSP-1NT had 4.2-fold increased odds of developing symptomatic P. vivax (PCR- and light microscopy-positive) compared with asymptomatic P. vivax malaria (PCR-positive and light microscopy-negative) (OR = 4.23, 95% CI 1.40 to 12.76, Figure 5).
Forest plot of the association of Pv MSP-1 N-terminus IgG responses with Plasmodium vivax outcomes. Estimates represent the estimate of P. vivax infection in IgG responders compared with non-responders. For cross-sectional studies, the estimate is an odds ratio; for cohort studies, it is a risk ratio. 1Symptomatic and asymptomatic P. vivax-positive individuals were compared with P. vivax-negative individuals; 2symptomatic individuals who were positive for P. vivax were compared with asymptomatic individuals who were either positive or negative for P. vivax; 3symptomatic individuals who were positive for P. vivax by both PCR and LM were compared with individuals who were negative for P. vivax by both PCR and LM. aEstimate calculated by the current authors from data in the paper; bdata supplied by the original authors and estimate calculated by the current authors; cpublished estimate. All estimates are unadjusted, with the exception of the estimate from Nogueira et al. [27], which was adjusted for geographical sector. When I 2 was ≥30%, meta-analysis based on a fixed-effects model was conducted. Abbreviations: LM, light microscopy; PNG, Papua New Guinea; W, weight.
Cohort studies
Meta-analysis of two cohort studies in Brazil showed that IgG responders to PvMSP-1NT had a 62% reduced risk of P. vivax infection compared with non-responders (feOR = 0.38, 95% CI 0.20 to 0.71, I 2 = 0%, Figure 5) [27],[31] indicating that PvMSP-1NT may be a target for protective immunity.
Association between antibody responses to PvMSP-3α and P. vivax
Members of the P. vivax MSP-3 multigene family, including PvMSP-3α, are structurally related to P. falciparum MSP-3, and are thought to associate with proteins anchored to the merozoite surface [53]. PvMSP-3α is highly polymorphic, with polymorphisms clustered in the N-terminal half of the central alanine-rich coiled-coil domain (Block I repeats, PvMSP-3αRI) and the less variable C-terminal half of the domain (Block II repeats, PvMSP-3αRII). By contrast, the extreme N-terminal (PvMSP-3αNT) and C-terminal (PvMSP-3αCT) domains are relatively conserved [54].
Cross-sectional studies
One cross-sectional study in Brazil by Lima-Junior et al.[25] and one cohort study in PNG by Stanisic et al.[36] (which contributed both cross-sectional and cohort data) investigated the association between antibody responses to regions of PvMSP-3α and P. vivax outcomes. The two cross-sectional studies gave opposing results and were not combined (I 2 > 75% and/or P < 0.01). Lima-Junior et al.[25] showed around a 60% reduction in the odds of P. vivax infection in IgG responders to PvMSP-3α full-length (MSP-3αFL), PvMSP-3αNT, PvMSP-3αRI, and around a 35% reduction for PvMSP-3αRII and PvMSP-3αCT compared with non-responders (Figure 6). Conversely, Stanisic et al.[36] showed fold increases of between 1.39 and 2.16 in the odds of P. vivax infection in PNG in IgG responders to PvMSP-3αNT, PvMSP-3αRI, PvMSP-3αRII, and PvMSP-3αCT, compared with non-responders (Figure 6).
Forest plot of the association of Pv MSP-3α IgG responses with Plasmodium vivax outcomes. Estimates represent the estimate of P. vivax infection in IgG responders compared with non-responders. For cross-sectional studies, estimate is an odds ratio; for cohort studies, it is a risk ratio. aData supplied by the original authors and estimate calculated by the current authors; bestimate calculated by the current authors from data in the paper; cpublished estimate. All estimates are unadjusted, with the exception of estimates from cohort data from Stanisic et al. [36], which were adjusted for age, season, spatial variation, and individual differences in exposure. Meta-analysis of responses to PvMSP-3αNT, PvMSP-3αRI, PvMSP-3αRII, and PvMSP-3αCT, and odds of P. vivax infection (estimates from cross-sectional studies) showed a high degree of heterogeneity (I 2 > 75% and/or P < 0.1) so estimates were not pooled. Abbreviations: LM, light microscopy; LDR-FMA, ligase detection reaction-fluorescent microsphere assay; PNG, Papua New Guinea.
Cohort studies
Although PvMSP-3α antibodies appeared to be a marker of P. vivax infection in cross-sectional data from Stanisic et al.[36], cohort data from the same study showed a 47% reduction in the risk of symptomatic P. vivax for PvMSP-3αRII IgG responders and around a 20% risk reduction for IgG responders to PvMSP-3αNT, PvMSP-3αRI and PvMSP-3αCT (Figure 6).
Association between antibody responses to PvMSP-5 and P. vivax
Case-control studies
The highly polymorphic PvMSP-5 contains potential signal and glycosylphosphatidyl inositol (GPI) anchor sequences and a single EGF-like domain near the carboxyl-terminus [55]. PvMSP-5 has been localized to the apical end of merozoites [56]. One case-control study in Indonesia showed no association between IgG (OR = 0.81, 95% CI 0.44 to 1.47) or IgM (OR = 1.12, 95% CI 0.62 to 2.04) responses to PvMSP-5 and odds of symptomatic P. vivax[33].
Association between antibody responses to PvMSP-9 and P. vivax
PvMSP-9 is associated with the surface of the merozoite and contains a hydrophobic signal sequence, a highly conserved N-terminal domain with a cluster of four cysteines, and a C-terminal region containing two species-specific blocks of repeats, designated PvMSP-9RI and PvMSP-9RII[57],[58]. Recombinant proteins may represent individual blocks or both blocks (PvMSP-9RIRII).
PvMSP-9 N-terminus
Cross-sectional studies
Meta-analysis of three sets of cross-sectional data [24],[26],[36] showed that IgG responses to PvMSP-9NT were associated with a 76% increase in odds of P. vivax infection compared with non-responders (pooled OR using random effects reOR = 1.76, 95% CI 0.95 to 3.25, I 2 = 48.7%, Figure 7) suggesting that PvMSP-9NT is a marker of exposure.
Forest plot of the association of Pv MSP-9 IgG responses with P. vivax outcomes. Estimates represent the estimate of P. vivax infection in IgG responders compared with non-responders. For cross-sectional studies, the estimate is an odds ratio, for cohort studies, it is a risk ratio. aData supplied by original authors and estimate calculated by the current authors; bestimate calculated by the current authors from data in the paper; cpublished estimate. All estimates are unadjusted, with the exception of estimates from cohort data from Stanisic et al. [36], which were adjusted for age, season, spatial variation, and individual differences in exposure. 1Meta-analysis of PvMSP-9RIIRII and PvMSP-9RII with odds of P. vivax infection showed a high degree of heterogeneity (I 2 = 77.5%, P = 0.012 and 87%, P = 0.006 respectively), so results were not pooled. Abbreviations: LDR-FMA, ligase detection reaction-fluorescent microsphere assay; LM, light microscopy; PNG, Papua New Guinea; W, weight.
Cohort studies
One cohort study by Stanisic et al.[36] also provided evidence for a protective effect of IgG responses to PvMSP-9NT, with a 40% reduction in the risk of symptomatic P. vivax (RR = 0.60, 95% CI 0.42 to 0.85).
PvMSP-9 block repeats
Cross-sectional studies
Three sets of cross-sectional data investigated the association between IgG antibodies to a protein representing the two blocks of repeats in PvMSP-9 (PvMSP-9RIRII) and P. vivax infection [24],[26],[36]. In a Brazilian study in 2012, Lima-Junior et al.[26] showed that IgG responders to PvMSP-9RIRII had a 67% reduction in the odds of P. vivax infection, compared with non-responders, but in a 2008 study by these authors [24] in the same region, PvMSP-9RIRII responders were found to have increased odds of P. vivax infection (OR = 2.64, Figure 7). Similar divergent results were also found in the 2008 study when IgG responses to PvMSP-9RII were examined [24]. Stanisic et al.[36] showed that IgG responders to PvMSP-9RIRII had a 33% increase in odds of P. vivax infection detected by LDR-FMA (OR = 1.33, 95% CI 0.50 to 3.53). Meta-analysis of PvMSP-9RIRII responses showed a high degree of heterogeneity in estimates (I2 = 77.5%, P = 0.012), and a pooled estimate was not reported.
Cohort studies
Cohort data from Stanisic et al.[36] showed no association between IgG responses to PvMSP-9RIRII and prospective risk of symptomatic P. vivax (RR = 0.97, 95% CI 0.51 to 1.82; Figure 7).
Association between antibody responses to PvAMA1 ectodomain and P. vivax
Cross-sectional and case-control studies
PvAMA1 is a type 1 transmembrane protein present in the microneme organelles of Plasmodium spp. A cross-sectional study in Turkey, which included both children and adults, investigated the association between anti-PvAMA1 ectodomain responses and prevalence of P. vivax infection, and found that total IgG responses (OR = 4.62, 95% CI 2.17 to 9.82, Figure 8) and IgM responses (OR = 2.22, 95% CI 1.06 to 4.67) were associated with increased odds of P. vivax infection [39]. A similar increase in the odds of P. vivax infection was seen in a nested case-control study [37] of pregnant women in Thailand (responders versus non-responders, OR = 4.25, 95% CI 2.08 to 8.70, Figure 8). PvAMA1 is therefore associated with P. vivax exposure in geographically and demographically diverse populations.
Forest plot of the association of Pv AMA1, Pv RBP1, and Pv SERA4 IgG responses with Plasmodium vivax infection. Estimates represent the odds of P. vivax infection in responders compared with non-responders. 1Colina study site; 2Ribeirinha study site. aEstimate supplied by original authors following correspondence; bdata supplied by original authors and estimate calculated by the current authors. All estimates are unadjusted, with the exception of the estimate from Fowkes et al. [37] which was adjusted for gravidity, trimester, and prophylaxis, and the estimates from Tran et al. [30], which were adjusted for age. When I 2 was ≥30%, meta-analysis based on a fixed-effects model was conducted. Abbreviations: LM, light microscopy; W, weight.
Association between antibody responses to PvRBP1 and P. vivax
Cross-sectional studies
PvRBP1 is a Type I integral membrane protein, which was identified based on its ability to adhere preferentially to reticulocyte-enriched populations of erythrocytes [59],[60]. Together with PvRBP2, it is thought to form a complex at the apical pole of the merozoite [59],[61]. Meta-analysis of two cross-sectional sets of data from two study sites by Tran et al.[30] indicated that IgG responders to PvRBP1 had a 76% increase in odds of P. vivax infection detected by light microscopy compared with non-responders (feOR = 1.76, 95% CI 0.55 to 5.59, I 2 = 0%, Figure 8).
Association between antibody responses to PvSERA4 and P. vivax
Cross-sectional studies
Serine repeat antigen 4 (SERA4) is the most dominantly expressed member of the P. vivax SERA multigene family, and its expression profile parallels that of PfSERA5, a blood-stage vaccine candidate [62]. A single cross-sectional study investigating the association between anti-PvSERA4 responses and P. vivax infection showed that both IgG and IgM responders had higher odds of P. vivax detected by light microscopy compared with non-responders (OR = 1.80, 95% CI 0.84 to 3.86, Figure 8 and OR = 2.50, 95% CI 1.39 to 4.49, respectively) [39].
Discussion
In this systematic review, we aimed to identify immunological biomarkers of P. vivax infection and protective immunity by standardizing estimates of the association between P. vivax antibodies and P. vivax outcomes across populations. We found a paucity of studies investigating associations between antibody responses to P. vivax antigens and risk of P. vivax, particularly cohort studies, and studies conducted in the Asia-Pacific [1]. Although there was considerable heterogeneity between studies, antibody responses to several antigens were associated with P. vivax infection and protective immunity to P. vivax. However, this review highlights the need for additional studies, and identifies several issues in the interpretation and reporting of data from epidemiological studies investigating immunity to P. vivax.
Studies included in the review represented diverse geographical populations living in areas of varying P. vivax endemicity. However, the geographical regions and countries represented were limited. Half of the studies provided data from the Asia-Pacific region, which represents 91% of the population at risk of P. vivax malaria [1], but only four countries were represented (Indonesia, Turkey, Thailand, and Papua New Guinea). The remaining half of the studies provided data from South America, representing only 6% of the population at risk of P. vivax malaria [1], but all were performed in Brazil. This predominance of data from Brazil has implications for the generalizability of findings to other P. vivax-endemic regions in South America and the Asia-Pacific. However, despite the population heterogeneity and the considerable heterogeneity in estimates observed, immunological markers of P. vivax infection could be identified: IgG responses to PvCSP, PvMSP-119, PvMSP-9NT, and PvAMA1 were associated with increased odds of P. vivax in geographically diverse populations. Other antigens were also shown to be markers of P. vivax infection, but only in single populations (PvMSP-3α, PvMSP-9RIRII, PvDBP, and PvRBP1). Serosurveillance using PvCSP in Korea [63]-[67] and PvMSP-119 and PvAMA1 in Vanuatu [68], Cambodia [69], and Somalia [70] has been employed to successfully map P. vivax transmission, and data from this review support their use in serosurveillance campaigns. However, this review highlights that further studies, conducted in diverse geographical settings and including additional antigens, are needed to ensure the generalizability of results across different populations with variable P. vivax transmission.
Protective immunity could only be examined in a handful of cohort studies, all of which showed evidence for protective blood-stage antibodies targeting PvMSP-119, PvMSP-1NT, PvMSP-3α and PvMSP-9NT antigens but only in single geographical locations. This was also the case for PvDBP, a prime vaccine target (because of its essential role in invasion) [44] that is currently in Phase I trials [5]. PvDBP was examined in only two cohort studies (which looked at different regions) and only Cole-Tobian et al.[34] showed evidence of allele-specific PvDBPII protective immunity against high-density parasitemia. Interestingly, no cohort study examined the protective effect of antibody responses to either the pre-erythrocytic antigen PvCSP or the gametocyte antigen Pvs25. Both of these have previously been assessed in Phase I trials [6],[71]-[73], and PfCSP comprises the current Phase III P. falciparum vaccine RTS,S, which has demonstrated around 50% efficacy in young children and around 30% efficacy in infants [15],[16]. This review shows that very few antigens meet the pre-clinical criteria for prioritizing candidate antigens (targets of protective immunity in humans) for vaccine development, which is particularly pertinent given the difficulties in meeting other in vitro pre-clinical criteria (demonstrating essential/important function, abundance, limited genetic diversity, inhibition of parasite growth, protection in animal models of infection) [7] because of difficulties in maintaining P. vivax in culture. In order to prioritize antigens for P. vivax vaccine development, further studies including additional antigens and established, clinically relevant end-points (for example, allele-specific responses with allele-specific end-points) are needed to provide valuable evidence for the role of particular P. vivax antigens in protective immunity.
The considerable heterogeneity observed in the estimates of association, which meant that the magnitude and the direction of effect estimates from different studies varied considerably, was a major issue in the meta-analyses, such that study estimates could not be reliably combined in some instances. Methodological diversity between studies may have contributed to the heterogeneity: antibody responses were measured in different ways (alleles, antigen preparation); P. vivax infection was determined using detection methods of varying sensitivities (PCR is more sensitive than light microscopy); and statistical methodology varied. Furthermore, the estimates from the majority of studies were unadjusted for potential confounders, and within-study bias may also have contributed to the heterogeneity observed. Transmission micro-epidemiology within study sites may be an important confounder, biasing the direction of effect in either way: individuals living in areas with the highest P. vivax exposure will acquire both biomarkers of exposure and protective immunity, but will also be at increased risk of future P. vivax infections. Study design may also be an important source of heterogeneity. The majority of studies were cross-sectional or case-control studies in which antibody responses and P. vivax outcomes had been determined at a single time point, in those with or without P. vivax outcome. Although we used this study design to identify immunological markers of P. vivax exposure, using data from a single time point has the potential to also capture a degree of protective immunity in the population. Indeed, we observed these types of divergent associations for several antigens, including studies by the same authors using the same methodology both in different populations [22] and within the same population [24],[26]. These findings highlight the limitations of using cross-sectional data, particularly when interpreting and comparing data across populations with varying degrees of P. vivax endemicity and immunity.
Differences in P. vivax transmission and exposure history will result in differential acquisition of immunity, which will influence associations between P. vivax antibody responses and clinical outcomes. To reduce bias in the systematic review, we excluded studies on transmigrants and studies in which the majority of the population resided in a malaria-endemic area for a short time. This bias was highlighted in two studies, which met the respective inclusion criteria, both by Lima-Junior et al. and performed in the same region of Brazil [24],[26]. PvMSP-9 IgG responders were found to have increased odds of P. vivax infection in 2008 [24], but decreased odds of infection in 2012 [26]. However, the population composition changed between the two studies: in 2008, 82% of participants were indigenous to the malaria-endemic area, compared with only 59% in 2012 (J. Ferreira, personal communication). This may explain, in part, the differences observed, because in both studies, time of residence in the malaria-endemic area was positively correlated with the anti-PvMSP-9 response [24],[26]. Differential effects according to transmission were also anecdotally observed: one study by Yidez-Zeyrek et al. in Turkey [39] showed greater magnitudes of effect with IgM than with IgG responses (PvMSP-119 and PvSERA4), indicating that individuals living in this P. vivax-endemic area had limited exposure to P. vivax. Interestingly, the ability of IgG to serve as a marker of exposure in this study was more than twice that of estimates from areas of higher P. vivax transmission (in Brazil and PNG), highlighting the potential for transmission intensity to influence results. Future studies should be aware of the potential confounding introduced by variations in P. vivax exposure and transmission intensity, particularly those conducted in areas in which P. vivax epidemiology is complicated by the presence of migrant workers or transient communities, which is common in P. vivax-endemic areas in South America and South-East Asia.
This review aimed to be as comprehensive as possible, and to identify all data by which an association between P. vivax responses and P. vivax outcomes could be examined. By contacting authors directly, we were able to obtain data from a further 11 studies for which data was not originally published. Commonly, these studies were descriptive in nature, comparing antibody prevalence in P. vivax infected versus uninfected individuals, with no quantification of the magnitude of effect. Consequently, many included studies were not sufficiently powered to detect a statistically significant association between antibody responses and P. vivax outcomes. Publication bias may also be an issue in the P. vivax immunity literature, which could not be assessed in this review because of the small number of studies in each analysis.
In this review, we also included total IgG subclasses, as well as IgG and IgM, to infer potential functional mechanisms, with similar associations seen with subclasses as to total IgG (see Additional file 5). IgG1 and IgG3 were the predominant subclasses to P. vivax antigens, and may function by opsonic phagocytosis [74], or by fixing complement. We found only one study that utilized a functional assay: King et al.[35] showed that binding inhibitory antibodies to PvDBPII were associated with protection from P. vivax infection. The lack of a continuous culturing system for P. vivax currently prohibits the use of most types of functional assays, but will clearly be important in future studies to determine the relative role of various immune mechanisms in protection against P. vivax.
Conclusion and future directions
In the absence of an in vitro system, population-based immunoepidemiology studies are pivotal to identify P. vivax antigens associated with protective immunity and exposure. This systematic review revealed antibody responses to several antigens that were associated with P. vivax infection and protective immunity. However, observations were often made in a small number of (sometimes single) studies, and further research is needed to validate these findings. More research is needed not only on P. vivax blood-stage antigens, but on sporozoite and gametocyte antigens, which are important markers of P. vivax transmission. Cohort studies are preferable, because they can be used to examine both markers of exposure and protective immunity. Future studies should aim to represent diverse populations, and special consideration in design and interpretation of findings should be given to studies in populations that contain considerable migrant sub-populations. Importantly, future studies should appropriately and comprehensively report data, and we have previously published guidelines to facilitate correct reporting of malaria immunoepidemiology observational studies (Proposed guidelines of the reporting of Malaria Immuno-epidemiology Observational Studies (MIOS guidelines) [11]). Additional well-reported studies, encompassing a wider geographical area, will provide a solid evidence base for P. vivax antigens in the use of vaccines and serosurveillance tools.
Authors’ contributions
JCC, RP, PAA, JAS, JGB, and FJIF designed the study and/or experiments. JCC, RP, PAA, JAS, JGB, and FJIF analyzed the data: JCC, RP, PAA, and FJIF collected data and/or performed experiments for the study. JCC and FJIF wrote the first draft of the paper JCC, RP, PAA, JAS, JGB, and FJIF contributed to the writing of the paper: All authors agree with the manuscript’s results and conclusions. All authors approved the final version of the manuscript.
Additional files
Abbreviations
- AMA1:
-
apical membrane antigen 1
- CI:
-
confidence interval
- CC:
-
case-control
- CS:
-
cross-sectional
- CSP:
-
circumsporozoite protein
- CT:
-
C-terminal
- DBP:
-
Duffy binding protein
- ELISA:
-
enzyme-linked immunosorbent assay
- FL:
-
full-length
- HR:
-
hazard ratio
- IRR:
-
incident rate ratio
- LDR-FMA:
-
ligase detection reaction-fluorescent microsphere assay
- LM:
-
light microscopy
- MSP:
-
merozoite surface protein
- NT:
-
N-terminal
- OR:
-
odds ratio
- Pf :
-
P. falciparum
- PNG:
-
Papua New Guinea
- Pv :
-
P. vivax
- RI:
-
Block I repeats
- RII:
-
Block II repeats
- RR:
-
risk ratio
- RBP1:
-
reticulocyte binding protein-1
- RTQ-PCR:
-
real-time quantitative polymerase chain reaction
- SERA4:
-
serine repeat antigen 4
References
Guerra CA, Howes RE, Patil AP, Gething PW, Van Boeckel TP, Temperley WH, Kabaria CW, Tatem AJ, Manh BH, Elyazar IR, Baird JK, Snow RW, Hay SI: The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl Trop Dis. 2010, 4: e774-10.1371/journal.pntd.0000774.
Anstey NM, Douglas NM, Poespoprodjo JR, Price RN: Plasmodium vivax: clinical spectrum, risk factors and pathogenesis. Adv Parasitol. 2012, 80: 151-201. 10.1016/B978-0-12-397900-1.00003-7.
A research agenda for malaria eradication: diagnoses and diagnostics. PLoS Med. 2011, 8: e1000396-10.1371/journal.pmed.1000396.
A research agenda for malaria eradication: vaccines. PLoS Med. 2011, 8: e1000398-10.1371/journal.pmed.1000398.
WHO: Malaria Vaccine Rainbow Tables. 2014. ., [http://www.who.int/vaccine_research/links/Rainbow/en/index.html]
Herrera S, Fernandez OL, Vera O, Cardenas W, Ramirez O, Palacios R, Chen-Mok M, Corradin G, Arevalo-Herrera M: Phase I safety and immunogenicity trial of Plasmodium vivax CS derived long synthetic peptides adjuvanted with montanide ISA 720 or montanide ISA 51. Am J Trop Med Hyg. 2011, 84: 12-20. 10.4269/ajtmh.2011.09-0516.
Richards JS, Beeson JG: The future for blood-stage vaccines against malaria. Immunol Cell Biol. 2009, 87: 377-390. 10.1038/icb.2009.27.
Mueller I, Galinski MR, Tsuboi T, Arevalo-Herrera M, Collins WE, King CL: Natural acquisition of immunity to Plasmodium vivax: epidemiological observations and potential targets. Adv Parasitol. 2013, 81: 77-131. 10.1016/B978-0-12-407826-0.00003-5.
Marsh K, Kinyanjui S: Immune effector mechanisms in malaria. Parasite Immunol. 2006, 28: 51-60. 10.1111/j.1365-3024.2006.00808.x.
White NJ: Determinants of relapse periodicity in Plasmodium vivax malaria. Malar J. 2011, 10: 297-10.1186/1475-2875-10-297.
Fowkes FJ, Richards JS, Simpson JA, Beeson JG: The relationship between anti-merozoite antibodies and incidence of Plasmodium falciparum malaria: a systematic review and meta-analysis. PLoS Med. 2010, 7: e1000218-10.1371/journal.pmed.1000218.
Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB: Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000, 283: 2008-2012. 10.1001/jama.283.15.2008.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P: Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009, 6: e1000097-10.1371/journal.pmed.1000097.
Zhang J, Yu KF: What’s the relative risk? A method of correcting the odds ratio in cohort studies of common outcomes. JAMA. 1998, 280: 1690-1691. 10.1001/jama.280.19.1690.
The RTS: S Clinical Trials Partnership: First results of phase 3 trial of RTS, S/AS01 malaria vaccine in African children. N Engl J Med. 2011, 365: 1863-1875. 10.1056/NEJMoa1102287.
The RTS: S Clinical Trials Partnership: A phase 3 trial of RTS, S/AS01 malaria vaccine in African infants. N Engl J Med. 2012, 367: 2284-2295. 10.1056/NEJMoa1208394.
DerSimonian R, Laird N: Meta-analysis in clinical trials. Control Clin Trials. 1986, 7: 177-188. 10.1016/0197-2456(86)90046-2.
Higgins JP, Thompson SG, Deeks JJ, Altman DG: Measuring inconsistency in meta-analyses. Br Med J. 2003, 327: 557-560. 10.1136/bmj.327.7414.557.
Ioannidis JP: Interpretation of tests of heterogeneity and bias in meta-analysis. J Eval Clin Pract. 2008, 14: 951-957. 10.1111/j.1365-2753.2008.00986.x.
Higgins JP, Thompson SG: Quantifying heterogeneity in a meta-analysis. Stat Med. 2002, 21: 1539-1558. 10.1002/sim.1186.
Deeks JJ HJ, Altman DG: Chapter 9: Analysing data and undertaking meta-analyses. In Cochrane Handbook for Systematic Reviews of Interventions, Version 5.1.0 (updated March 2011). Edited by Higgins JPT, Green S. The Cochrane Collaboration; 2011. Available from ., [www.cochrane-handbook.org]
Fernandez-Becerra C, Sanz S, Brucet M, Stanisic DI, Alves FP, Camargo EP, Alonso PL, Mueller I, del Portillo HA: Naturally-acquired humoral immune responses against the N- and C-termini of the Plasmodium vivax MSP1 protein in endemic regions of Brazil and Papua New Guinea using a multiplex assay. Malar J. 2010, 9: 29-10.1186/1475-2875-9-29.
Kano FS, Sanchez BA, Sousa TN, Tang ML, Saliba J, Oliveira FM, Nogueira PA, Goncalves AQ, Fontes CJ, Soares IS, Brito CF, Rocha RS, Carvalho LH: Plasmodium vivax Duffy binding protein: baseline antibody responses and parasite polymorphisms in a well-consolidated settlement of the Amazon region. Trop Med Int Health. 2012, 17: 989-1000. 10.1111/j.1365-3156.2012.03016.x.
Lima-Junior JC, Tran TM, Meyer EV, Singh B, De-Simone SG, Santos F, Daniel-Ribeiro CT, Moreno A, Barnwell JW, Galinski MR, Oliveira-Ferreira J: Naturally acquired humoral and cellular immune responses to Plasmodium vivax merozoite surface protein 9 in Northwestern Amazon individuals. Vaccine. 2008, 26: 6645-6654. 10.1016/j.vaccine.2008.09.029.
Lima-Junior JC, Jiang J, Rodrigues-da-Silva RN, Banic DM, Tran TM, Ribeiro RY, Meyer VSE, De-Simone SG, Santos F, Moreno A, Barnwell JW, Galinski MR, Oliveira-Ferreira J: B cell epitope mapping and characterization of naturally acquired antibodies to the Plasmodium vivax merozoite surface protein-3α (PvMSP-3 α) in malaria exposed individuals from Brazilian Amazon. Vaccine. 2011, 29: 1801-1811. 10.1016/j.vaccine.2010.12.099.
Lima-Junior JC, Rodrigues-da-Silva RN, Banic DM, Jiang J, Singh B, Fabricio-Silva GM, Porto LC, Meyer EV, Moreno A, Rodrigues MM, Barnwell JW, Galinski MR, de Oliveira-Ferreira J: Influence of HLA-DRB1 and HLA-DQB1 alleles on IgG antibody response to the P. vivax MSP-1, MSP-3alpha and MSP-9 in individuals from Brazilian endemic area. PLoS One. 2012, 7: e36419-10.1371/journal.pone.0036419.
Nogueira PA, Alves FP, Fernandez-Becerra C, Pein O, Santos NR, da Silva LHP, Camargo EP, del Portillo HA: A reduced risk of infection with Plasmodium vivax and clinical protection against malaria are associated with antibodies against the N terminus but not the C terminus of merozoite surface protein 1. Infect Immun. 2006, 74: 2726-2733. 10.1128/IAI.74.5.2726-2733.2006.
Oliveira-Ferreira J, Pratt-Riccio LR, Arruda M, Santos F, Ribeiro CT, Goldberg AC, Banic DM: HLA class II and antibody responses to circumsporozoite protein repeats of P. vivax (VK210, VK247 and P. vivax-like) in individuals naturally exposed to malaria. Acta Trop. 2004, 92: 63-69. 10.1016/j.actatropica.2004.02.011.
Souza-Silva FA, Da Silva-Nunes M, Sanchez BAM, Ceravolo IP, Malafronte RS, Brito CFA, Ferreira MU, Carvalho LH: Naturally acquired antibodies to Plasmodium vivax Duffy binding protein (DBP) in rural Brazilian Amazon. Am J Trop Med Hyg. 2010, 82: 185-193. 10.4269/ajtmh.2010.08-0580.
Tran TM, Oliveira-Ferreira J, Moreno A, Santos F, Yazdani SS, Chitnis CE, Altman JD, Meyer EV, Barnwell JW, Galinski MR: Comparison of IgG reactivities to Plasmodium vivax merozoite invasion antigens in a Brazilian Amazon population. Am J Trop Med Hyg. 2005, 73: 244-255.
Versiani FG, Almeida ME, Melo GC, Versiani FO, Orlandi PP, Mariuba LA, Soares LA, Souza LP, da Silva Balieiro AA, Monteiro WM, Costa FT, del Portillo HA, Lacerda MV, Nogueira PA: High levels of IgG3 anti ICB2-5 in Plasmodium vivax-infected individuals who did not develop symptoms. Malar J. 2013, 12: 294-10.1186/1475-2875-12-294.
Ak M, Jones TR, Charoenvit Y, Kumar S, Kaslow DC, Maris D, Marwoto H, Masbar S, Hoffman SL: Humoral immune responses against Plasmodium vivax MSP1 in humans living in a malaria endemic area in Flores, Indonesia. Southeast Asian J Trop Med Public Health. 1998, 29: 685-691.
Woodberry T, Minigo G, Piera KA, Hanley JC, de Silva HD, Salwati E, Kenangalem E, Tjitra E, Coppel RL, Price RN, Anstey NM, Plebanski M: Antibodies to Plasmodium falciparum and Plasmodium vivax merozoite surface protein 5 in Indonesia: species-specific and cross-reactive responses. J Infect Dis. 2008, 198: 134-142. 10.1086/588711.
Cole-Tobian JL, Michon P, Biasor M, Richards JS, Beeson JG, Mueller I, King CL: Strain-specific duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous Plasmodium vivax strains in Papua New Guinean children. Infect Immun. 2009, 77: 4009-4017. 10.1128/IAI.00158-09.
King CL, Michon P, Shakri AR, Marcotty A, Stanisic D, Zimmerman PA, Cole-Tobian JL, Mueller I, Chitnis CE: Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci U S A. 2008, 105: 8363-8368. 10.1073/pnas.0800371105.
Stanisic DI, Javati S, Kiniboro B, Lin E, Jiang J, Singh B, Meyer EV, Siba P, Koepfli C, Felger I, Galinski MR, Mueller I: Naturally Acquired immune responses to P. vivax merozoite surface protein 3alpha and merozoite surface protein 9 are associated with reduced risk of P. vivax malaria in young Papua New Guinean children. PLoS Negl Trop Dis. 2013, 7: e2498-10.1371/journal.pntd.0002498.
Fowkes FJ, McGready R, Cross NJ, Hommel M, Simpson JA, Elliott SR, Richards JS, Lackovic K, Viladpai-Nguen J, Narum D, Tsuboi T, Anders RF, Nosten F, Beeson JG: New insights into acquisition, boosting, and longevity of immunity to malaria in pregnant women. J Infect Dis. 2012, 206: 1612-1621. 10.1093/infdis/jis566.
Wongsrichanalai C, Webster HK, Permpanich B, Chuanak N, Ketrangsri S: Naturally acquired circumsporozoite antibodies and their role in protection in endemic falciparum and vivax malaria. Am J Trop Med Hyg. 1991, 44: 201-204.
Yildiz Zeyrek F, Palacpac N, Yuksel F, Yagi M, Honjo K, Fujita Y, Arisue N, Takeo S, Tanabe K, Horii T, Tsuboi T, Ishii KJ, Coban C: Serologic markers in relation to parasite exposure history help to estimate transmission dynamics of Plasmodium vivax. PLoS One. 2011, 6: e28126-10.1371/journal.pone.0028126.
Yildiz Zeyrek F, Babaoglu A, Demirel S, Erdogan DD, Ak M, Korkmaz M, Coban C: Analysis of naturally acquired antibody responses to the 19-kd C-terminal region of merozoite surface protein-1 of Plasmodium vivax from individuals in Sanliurfa. Turkey Am J Trop Med Hyg. 2008, 78: 729-732.
Nardin EH, Nussenzweig V, Nussenzweig RS, Collins WE, Harinasuta KT, Tapchaisri P, Chomcharn Y: Circumsporozoite proteins of human malaria parasites Plasmodium falciparum and Plasmodium vivax. J Exp Med. 1982, 156: 20-30. 10.1084/jem.156.1.20.
Rosenberg R, Wirtz RA, Lanar DE, Sattabongkot J, Hall T, Waters AP, Prasittisuk C: Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989, 245: 973-976. 10.1126/science.2672336.
Qari SH, Shi Y-P, Goldman IF, Alpers MP, Collins WE, Lal AA: Identification of Plasmodium vivax-like human malaria parasite. Lancet. 1993, 341: 780-783. 10.1016/0140-6736(93)90559-Y.
Beeson JG, Crabb BS: Towards a vaccine against Plasmodium vivax malaria. PLoS Med. 2007, 4: e350-10.1371/journal.pmed.0040350.
Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH: The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med. 1996, 184: 1531-1536. 10.1084/jem.184.4.1531.
Ranjan A, Chitnis CE: Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc Natl Acad Sci U S A. 1999, 96: 14067-14072. 10.1073/pnas.96.24.14067.
Fraser T, Michon P, Barnwell JW, Noe AR, Al-Yaman F, Kaslow DC, Adams JH: Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect Immun. 1997, 65: 2772-2777.
Tanabe K, Escalante A, Sakihama N, Honda M, Arisue N, Horii T, Culleton R, Hayakawa T, Hashimoto T, Longacre S, Pathirana S, Handunnetti S, Kishino H: Recent independent evolution of msp1 polymorphism in Plasmodium vivax and related simian malaria parasites. Mol Biochem Parasitol. 2007, 156: 74-79. 10.1016/j.molbiopara.2007.07.002.
Pasay MC, Cheng Q, Rzepczyk C, Saul A: Dimorphism of the C terminus of the Plasmodium vivax merozoite surface protein 1. Mol Biochem Parasitol. 1995, 70: 217-219. 10.1016/0166-6851(95)00015-S.
Holder AA, Blackman MJ, Burghaus PA, Chappel JA, Ling IT, McCallum-Deighton N, Shai S: A malaria merozoite surface protein (MSP1)-structure, processing and function. Mem Inst Oswaldo Cruz. 1992, 87: 37-42. 10.1590/S0074-02761992000700004.
Blackman MJ, Heidrich HG, Donachie S, McBride JS, Holder AA: A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J Exp Med. 1990, 172: 379-382. 10.1084/jem.172.1.379.
Blackman MJ, Ling IT, Nicholls SC, Holder AA: Proteolytic processing of the Plasmodium falciparum merozoite surface protein-1 produces a membrane-bound fragment containing two epidermal growth factor-like domains. Mol Biochem Parasitol. 1991, 49: 29-33. 10.1016/0166-6851(91)90127-R.
Galinski MR, Corredor-Medina C, Povoa M, Crosby J, Ingravallo P, Barnwell JW: Plasmodium vivax merozoite surface protein-3 contains coiled-coil motifs in an alanine-rich central domain. Mol Biochem Parasitol. 1999, 101: 131-147. 10.1016/S0166-6851(99)00063-8.
Rayner JC, Corredor V, Feldman D, Ingravallo P, Iderabdullah F, Galinski MR, Barnwell JW: Extensive polymorphism in the Plasmodium vivax merozoite surface coat protein MSP-3alpha is limited to specific domains. Parasitology. 2002, 125: 393-405. 10.1017/S0031182002002317.
Gomez A, Suarez CF, Martinez P, Saravia C, Patarroyo MA: High polymorphism in Plasmodium vivax merozoite surface protein-5 (MSP5). Parasitology. 2006, 133: 661-672. 10.1017/S0031182006001168.
Black CG, Barnwell JW, Huber CS, Galinski MR, Coppel RL: The Plasmodium vivax homologues of merozoite surface proteins 4 and 5 from Plasmodium falciparum are expressed at different locations in the merozoite. Mol Biochem Parasitol. 2002, 120: 215-224. 10.1016/S0166-6851(01)00458-3.
Vargas-Serrato E, Barnwell JW, Ingravallo P, Perler FB, Galinski MR: Merozoite surface protein-9 of Plasmodium vivax and related simian malaria parasites is orthologous to p101/ABRA of P. falciparum. Mol Biochem Parasitol. 2002, 120: 41-52. 10.1016/S0166-6851(01)00433-9.
Barnwell JW, Galinski MR, DeSimone SG, Perler F, Ingravallo P: Plasmodium vivax, P. cynomolgi, and P. knowlesi: identification of homologue proteins associated with the surface of merozoites. Exp Parasitol. 1999, 91: 238-249. 10.1006/expr.1998.4372.
Galinski MR, Medina CC, Ingravallo P, Barnwell JW: A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992, 69: 1213-1226. 10.1016/0092-8674(92)90642-P.
Galinski MR, Xu M, Barnwell JW: Plasmodium vivax reticulocyte binding protein-2 (PvRBP-2) shares structural features with PvRBP-1 and the Plasmodium yoelii 235 kDa rhoptry protein family. Mol Biochem Parasitol. 2000, 108: 257-262. 10.1016/S0166-6851(00)00219-X.
Galinski MR, Barnwell JW: Plasmodium vivax: merozoites, invasion of reticulocytes and considerations for malaria vaccine development. Parasitology Today (Personal ed). 1996, 12: 20-29. 10.1016/0169-4758(96)80641-7.
Palacpac NM, Leung BW, Arisue N, Tanabe K, Sattabongkot J, Tsuboi T, Torii M, Udomsangpetch R, Horii T: Plasmodium vivax serine repeat antigen (SERA) multigene family exhibits similar expression patterns in independent infections. Mol Biochem Parasitol. 2006, 150: 353-358. 10.1016/j.molbiopara.2006.07.006.
Cho D, Kim KH, Park SC, Kim YK, Lee KN, Lim CS: Evaluation of rapid immunocapture assays for diagnosis of Plasmodium vivax in Korea. Parasitol Res. 2001, 87: 445-448. 10.1007/s004360000360.
Kim S, Ahn HJ, Kim TS, Nam HW: ELISA detection of vivax malaria with recombinant multiple stage-specific antigens and its application to survey of residents in endemic areas. Korean J Parasitol. 2003, 41: 203-207. 10.3347/kjp.2003.41.4.203.
Lee KN, Suh IB, Chang EA, Kim SD, Cho NS, Park PW, An SS, Park O, Lim C: Prevalence of antibodies to the circumsporozite protein of Plasmodium vivax in five different regions of Korea. Trop Med Int Health. 2003, 8: 1062-1067. 10.1046/j.1360-2276.2003.01136.x.
Park SK, Lee KW, Hong SH, Kim DS, Lee JH, Jeon BH, Kim WS, Shin HJ, An SH, Park H: Development and evaluation of an immunochromatographic kit for the detection of antibody to Plasmodium vivax infection in South Korea. Yonsei Med J. 2003, 44: 747-750.
Suh IB, Lee KH, Kim YR, Woo SK, Kang HY, Won YD, An SS, Cho M, Cho D, Lim CS: Comparison of immunological responses to the various types circumsporozoite proteins of Plasmodium vivax in malaria patients of Korea. Microbiol Immunol. 2004, 48: 119-123. 10.1111/j.1348-0421.2004.tb03497.x.
Cook J, Reid H, Iavro J, Kuwahata M, Taleo G, Clements A, McCarthy J, Vallely A, Drakeley C: Using serological measures to monitor changes in malaria transmission in Vanuatu. Malar J. 2010, 9: 169-10.1186/1475-2875-9-169.
Cook J, Speybroeck N, Sochanta T, Somony H, Sokny M, Claes F, Lemmens K, Theisen M, Soares IS, D’Alessandro U, Coosemans M, Erhart A: Sero-epidemiological evaluation of changes in Plasmodium falciparum and Plasmodium vivax transmission patterns over the rainy season in Cambodia. Malar J. 2012, 11: 86-10.1186/1475-2875-11-86.
Bousema T, Youssef RM, Cook J, Cox J, Alegana VA, Amran J, Noor AM, Snow RW, Drakeley C: Serologic markers for detecting malaria in areas of low endemicity, Somalia, 2008. Emerg Infect Dis. 2010, 16: 392-399. 10.3201/eid1603.090732.
Schwartz L, Brown GV, Genton B, Moorthy VS: A review of malaria vaccine clinical projects based on the WHO rainbow table. Malaria J. 2012, 11: 11-10.1186/1475-2875-11-11.
Malkin EM, Durbin AP, Diemert DJ, Sattabongkot J, Wu Y, Miura K, Long CA, Lambert L, Miles AP, Wang J, Stowers A, Miller LH, Saul A: Phase 1 vaccine trial of Pvs25H: a transmission blocking vaccine for Plasmodium vivax malaria. Vaccine. 2005, 23: 3131-3138. 10.1016/j.vaccine.2004.12.019.
Wu Y, Ellis RD, Shaffer D, Fontes E, Malkin EM, Mahanty S, Fay MP, Narum D, Rausch K, Miles AP, Aebig J, Orcutt A, Muratova O, Song G, Lambert L, Zhu D, Miura K, Long C, Saul A, Miller LH, Durbin AP: Phase 1 trial of malaria transmission blocking vaccine candidates Pfs25 and Pvs25 formulated with montanide ISA 51. PLoS One. 2008, 3: e2636-10.1371/journal.pone.0002636.
Bredius RG, Fijen CA, De Haas M, Kuijper EJ, Weening RS, Van de Winkel JG, Out TA: Role of neutrophil Fc gamma RIIa (CD32) and Fc gamma RIIIb (CD16) polymorphic forms in phagocytosis of human IgG1- and IgG3-opsonized bacteria and erythrocytes. Immunology. 1994, 83: 624-630.
Acknowledgments
We thank Robyn Damary-Homan and Cameron McPherson for pilot literature searches and data extraction. We also thank John Adams, Mercia Arruda, Sukla Biswas, Tom Burkot, Luzia Carvalho, Cevayir Coban, Giuseppe Del Giudice, Yagya Dutta Sharma, Annette Erhart, Marcelo Ferreira, Daniel Gordon, Patricia Graves, Stephen Hoffman, Trevor Jones, Flora Kano, Chris King, Peter Kremsner, Kee-Hyoung Lee, George Lewis, Josue Lima-Junior, Ivo Mueller, Paulo Noguira, Ruth Nussenzweig, Joseli de Oliveira-Ferreira, Danielle Stanisic, Tuan Tran, Eileen Villasante, Chansuda Wongsrichanalai, and Sedigheh Zakeri for providing information and/or data for the systematic review.
Financial disclosure
This work was supported by the National Health and Medical Research Council of Australia (project grant, fellowships to FJIF and JGB, and an Infrastructure for Research Institutes Support Scheme Grant), the Australian Research Council (Future Fellowship to FJIF and JGB), and a Victorian State Government Operational Infrastructure Support grant. The funders had no role in study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that no competing interests exist.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Cutts, J.C., Powell, R., Agius, P.A. et al. Immunological markers of Plasmodium vivaxexposure and immunity: a systematic review and meta-analysis. BMC Med 12, 150 (2014). https://doi.org/10.1186/s12916-014-0150-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12916-014-0150-1