Abstract
Colorectal cancer (CRC) is a gastrointestinal tumor that develops from the colon, rectum, or appendix. The prognosis of CRC patients especially those with metastatic lesions remains unsatisfactory. Although various conventional methods have been used for the treatment of patients with CRC, the early detection and identification of molecular mechanisms associated with CRC is necessary. The scientific literature reports that altered expression of long non-coding RNAs (lncRNAs) contributed to the pathogenesis of CRC cells. LncRNA TUG1 was reported to target various miRNAs and signaling pathways to mediate CRC cell proliferation, migration, and metastasis. Therefore, TUG1 might be a potent predictive/prognostic biomarker for diagnosis of CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a gastrointestinal malignancy, ranked as the third most commonly diagnosed, and the second cause of cancer mortality worldwide [1]. This type of cancer originates from the colon, rectum, or appendix [2]. Pieces of evidence showed that the CRC incidence rate has risen during the past decades, specifically in developing countries [3]. Over 1.85 million new CRC cases are reported annually, with an increasing number of young people before the age of 50 [4]. Several factors such as genetics, epigenetics, and environment distributed across the CRC etiology and are responsible for disease heterogeneity [5, 6]. Etiologically, the three patterns of disease onset are sporadic, familial, and hereditary forms that affected 70%, 25%, and 5% of patients [7, 8]. It has been found that the age, environmental factors, dietary, lifestyle, gut microbiota, and genetic changes predispose persons to CRC [9]. Current treatment of CRC in primary and metastatic patients include laparoscopic surgery for primary disease, resection in case of metastatic tumors, radiotherapy for rectal neoplasm along with palliative, and neoadjuvant chemotherapies [10]. Besides, antibodies, probiotics, agarose tumor macrobeads, and gold-based drugs or their combinations are used for patients with CRC [11]. It has been found that the combination of chemotherapy and anti-EGFR (epidermal growth factor receptor) monoclonal antibodies cetuximab and panitumumab can prolong the median survival rate of these patients by 2 to 4 months compared with chemotherapy alone [12]. However, the impact of these therapies on the 5-year survival remains limited and very expensive [13, 14]. Several mutations in oncogenes, tumor suppressor genes, and genes associated with DNA repair have been identified as genetic risk factors for CRCs [15]. It has been reported that genetic mutations in SMAD4, BRAF, KRAS, PIK3CA, SMAD2, PTEN, and c-MYC play essential roles in patients with CRC [11]. Recent studies demonstrated that long non-coding RNAs (lncRNAs) as a subgroup of RNAs longer than 200 nucleotides presented important functions in the pathogenesis of CRC [16,17,18]. Aberrant expression of lncRNA is associated with several diseases, as well [17]. Previous investigations reported that lncRNA TUG1 showed tumor-suppressive or oncogenic functions in different types of cancers [19, 20]. Studies revealed that TUG1 expression was increased in CRC tumor tissues and promoted cell proliferation [21,22,23]. Further analysis revealed a significant negative correlation between the levels of TUG1 and the overall survival rate of patients with CRC [24]. In this review, we summarized functional roles of this lncRNA in the tumorigenesis of CRC cells.
Biological properties of lncRNA TUG1
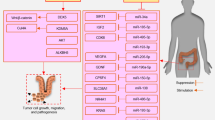
An initial genomic screening for genes upregulated in response to taurine treatment in developing mouse retinal cells detected taurine-upregulated gene 1 (TUG1) (also known as TI-227 H, LINC00080, and NCRNA00080), a 7.1-kb lncRNA that in the human genome is located on chromosome 22q12.2 [25]. Functional studies revealed mice lacking TUG1 had impaired retinal development [26]. There is also evidence that TUG1 is transcriptionally regulated by p53 [27]. The polycomb-repressive complex 2 (PRC2) contains enhancer of zeste homologue 2 (EZH2), suppressor of zeste 12 (SUZ12), and embryonic ectoderm development (EED) [28] that catalyzes lysine residue 27 di- and trimethylation on histone 3 (H3K27me3) to repress gene expression [29]. TUG1 by recruiting and binding to the PRC2 complex functions as a dynamic scaffold [30, 31]. TUG1 knockdown induced an upregulation of the cell-cycle genes, suggesting that TUG1 is involved in both cell proliferation and apoptosis [22]. TUG1 as a potent epigenetic regulator can mediate histone modification and DNA methylation in target genes [24, 32]. Besides, TUG1 by acting as a competing endogenous RNA (ceRNA) could sponge and repress microRNA (miRNA) [33]. Figure 1 displays different functions of TUG1.
TUG1 has recently been proposed as an oncogene in several types of cancer [34,35,36]., TUG1 is associated with large tumor size, advanced pathological stages, and distant metastasis [37, 38]. Experimental studies have disclosed that TUG1 significantly stimulated tumor cell proliferation, invasion, colony formation, and drug resistance in CRC cells (Table 1). Overexpression of TUG1 via mediating epithelial-mesenchymal transition (EMT)-associated gene expression, reduction of E-cadherin expression, and boosted the vimentin, N cadherin, and fibronectin expression promoted the invasion and metastasis of CRC cells [24, 39, 40].
The critical TUG1/miRNAs/transcription factors axes in colorectal cancer
There is growing evidence that TUG1 by targeting several signaling pathways plays critical roles in the progression of CRC cells [24]. Here, we described multiple miRNAs/transcription factors axes (Fig. 2) that can be regulated via this lncRNA in CRC.
TUG1/miR-145-5p/TRPC6
High expression of TUG1 has been proved to be correlated with CRC pathogenesis including proliferative along with the migratory ability, cell viability, tumor growth, and subcutaneous tumor formation [41, 42]. TUG1 is known to interact with miR-145-5p and regulate CRC cellular processes. Moreover, miR-145-5p suppressed the expression transient receptor potential cation channel subfamily c member 6 (TRPC6) as its candidate target, which its overexpression brought back miR-145-5p function in CRC. Altogether, TUG1 induces progression of CRC through miR-145-5p/TRPC6 axis, thereby regarding as a possible diagnostic marker for CRC management [42].
TUG1/miR-138-5p/ZEB2
It has been shown that high expression of TUG1 is correlated with increased proliferation and invasion along with reduced apoptosis of CRC cells [43]. TUG1 overexpression was also closely associated with the overall survival of CRC patients, indicating TUG1 as poor prognosis biomarker for CRC [44]. Zinc finger E-box binding homeobox 2 (ZEB2) is a binding protein that participated in CRC metastasis and associated with human CRC poor prognosis [45]. There was a positive and negative correlation between TUG1 with ZEB2 and miR-138-5p, respectively. Low expression of ZEB2 or overexpression of miR-138-5p reversed the induction of EMT which was caused by TUG1 overexpression. Therefore, TUG1 by suppressing the miR-138-5p/ZEB2 axis facilitated CRC occurrence and metastasis [3, 41, 43, 44].
TUG1/Wnt/β-catenin
High expression of TUG1 has been implicated in clinicopathological features of CRC including advanced tumor stage along with reduced overall survival and disease-free survival [24, 44, 46]. The Wnt/β-catenin signaling was found to transcriptionally regulate CRC proliferation [47]. TUG1 silencing resulted in low activity of Wnt/β-catenin and suppression of proliferation. In the CRC xenograft model, low expression of TUG1 inhibited both tumorigenicity and β-catenin nuclear localization. TUG1 through changing the nuclear localization of β-catenin reduced the Wnt/β-catenin signaling activity and subsequent induced CRC proliferation. Therefore, TUG1/Wnt/β-catenin could exert promising advancement for CRC prevention [44, 46].
TUG1/TGF-β/TWIST1
Transforming growth factor-beta (TGF-β) is involved in CRC tumorigenesis [48]. A recent study reported that TGF-β induced migration of CRC and overexpressed TUG1 as its downstream molecule. Recent findings demonstrated that TUG1 blockade suppressed migration, invasion, in vitro EMT along with in vivo lung metastasis. Hence, TUG1 is necessary for TGF-β-promoted pathophysiological features of CRC [49]. Twist family BHLH transcription factor 1 (TWIST1) stands as a kind of transcriptional modulator which is activated by TGF-β, resulting in low expression of E-cadherin [50]. TWIST1 silencing using siRNA resulted in significant reduction of CRC migration and EMT. It can be concluded that TGF-β-induced metastasis of CRC is regulated through the TUG1/TWIST1/EMT network, highlighting TUG1 as a novel target to inactivate the TGF-β signaling [49].
TUG1/miR-421/KDM2A/ERK/SP1
Specificity protein 1 (SP1) has a positive-regulated manner with TUG1 in CRC cells [51]. SP1 as an oncogene was found to promote CRC progression and metastasis [51, 52]. TUG1 loss of function inhibited cell growth and induced apoptosis of CRC cells [51]. Growing evidence revealed a negative correlation between TUG1 and miR-421 as a CRC tumor suppressor factor [53, 54]. Lysine demethylase 2 A (KDM2A) is a CRC oncogenic gene and a target for miR-421. TUG1 by sponging miR-421 induced KDM2A expression [51, 55]. Moreover, TUG1 has been found to intensify in vitro progression of CRC through the ERK signaling. Also, SP1 promoted in vivo CRC tumorigenesis by miR-421 suppression and KDM2A induction through TUG1 overexpression. Altogether, TUG1 as an oncogene can interact with SP1 and the miR-421/ KDM2A/ERK axis to facilitate CRC progression [51].
TUG1/miR-542-3p/TRIB2/Wnt/β-catenin
Tribbles homolog 2 (TRIB2) is an atypical protein kinase that has been dramatically upregulated with TUG1 in CRC tissues and cells [56, 57]. High expression of TUG1 by suppressing miR-542-3p was associated with tumor stage, lymph node metastasis, and histological differentiation of CRC patients. TUG1 or TRIB2 loss of function prohibited proliferation, migration, invasion along with in vivo tumor growth but facilitated CRC apoptosis. Besides, upregulation of TRIB2 as a miR-542-3p target reversed the impact of TUG1 silencing on CRC progression [58]. Considering the role of the Wnt/β-catenin signaling in CRC development, miR-542-3p suppression or TRIB2 upregulation has been reported to partly bring back the inhibitory function of TUG1 knockdown on the Wnt/β-catenin signaling. ThereforeTUG1 is regarded as a tumor promoter that stimulated CRC pathogenesis and drug resistance through the miR-542-3p/TRIB2 axis [58, 59].
TUG1/miR-153-1/KLF4
In contrast to highly-expressed TUG1 in CRC, miR-153-1 was under expressed. Depletion of TUG1 using si-TUG1 as well as ectopic expression of miR-153-1 repressed the proliferative and migratory capacity of CRC cells. Also, upregulated TUG1 reversed miR-153-1-mediated suppression of CRC cells [60]. Kruppel-like factor 4 (KLF4) is a zinc finger transcription factor that plays as a tumor suppressive gene in CRC [61]. Recent findings identified KLF4 as a direct transcription factor for miR-153-1 can suppress CRC pathogenesis but its expression negatively modulated by TUG1. Interestingly, TUG1-deficient mice demonstrated high and low expressions of E-cadherin along with N-cadherin as tumor metastasis-correlated EMT markers exerting the TUG1/miR-153-1/KLF4 axis in in vivo EMT of CRC cells. Such regulatory axis might provide great insights for either diagnostic or treatment possibility of CRC [60].
Chemoresistance features of TUG1
Chemotherapy in combination with targeted therapy has been found to impair tumor recurrence and increase survival rate of CRC patients, but chemotherapeutic resistance is considered as the leading cause of CRC therapy failure.Therefore, molecular knowledge of chemotherapeutic resistance is required for CRC tumor biology [62, 63]. 5-fluorouracil (5-FU) is regarded as an effective first-line drug for CRC patients, but unknown molecular approaches are still complicated its recovery features [17, 64]. Accumulating data demonstrated that TUG1 is overexpressed in 5-FU resistant CRC tissue and cells, which were associated with poor prognosis. TUG1 blockade was implicated to dramatically promote CRC cells sensitive to 5-FU through suppressing CRC cell apoptosis which is regulated by miR-197-3p and TYMS as a direct target of miR-197-3p. Such findings highlighted the possible significance of TUG1 as a predictive marker for exerting CRC response to 5-FU therapy and indicated TUG1 silencing as a novel therapeutic approach to reverse 5-FU resistance [65]. Besides, cancer stem cells are shown to be involved in CRC chemoresistance [66]. It has been found that TUG1 silencing inhibited CRC stem cell resistant to oxaliplatin through reducing GATA6 and targeting the BMP pathway. Altogether, TUG1 promoted CRC stem cell features and chemotherapeutic resistance via inducing the stability of the GATA6 protein, providing promising insights for CRC clinical therapy [67]. Improvement of drug resistant sensitivity remains an immediate necessity for CRC chemotherapies. CRC resistance to methotrexate (MTX) as the earliest cytotoxic drugs is still a main challenge to the physicians [62, 68]. A recent study indicated that TUG1 repressed CRC cell sensitivity to MTX through targeting its interaction with miR-186. TUG1 blockade re-sensitized CRC cell resistant to MTX. Indeed, a negative correlation between miR-186 and the cytoplasmic polyadenylation element binding protein 2 (CPEB2) protein has been shown in MTX resistant tumors. Therefore, TUG1 regulated CRC resistant to MTX through targeting miR-186 and consequent induction of CPEB2 expression, thereby holding TUG1 as a possible target for CRC management [69].
Insulin-like growth factor-2 mRNA–binding protein (IGF2BP) family members as a kind of RNA-binding proteins participated in tumorigenesis as well as chemoresistance via influencing either stability, translatability, or localization of lncRNA [70,71,72].IIt has been found that TUG1 and IGF2BP2 were both high-expressed in CRC cell resistant to cisplatin through autophagy activation. Low TUG1 expression decreased CRC chemoresistance to cisplatin and facilitated miR-195-5p expression. Therefore, the TUG1/IGF2BP2/miR-195-5p axis intensify CRC cell growth and induce such malignant cell resistance to cisplatin, regarding as underlying target for CRC therapy [73].
Prognostic significance of TUG1 in colorectal cancer
Along with the biological behaviors of TUG1 in regulating CRC pathogenesis, it is also emerging as a crucial substrate for the progress of CRC biomarkers for early detection, prognosis prediction, and anticipating therapy response to diverse chemotherapies and developing therapies [74]. Currently, a study proposed that TUG1 could play a vital function in CRC metastasis. following investigation of the TUG1 expression levels in 120 CRC patients, high TUG1 expression was observed in tumor tissue which was closely associated with the poor survival time of the CRC patients [24, 75, 76]. Further in vitro experiments revealed the oncogenic impact of TUG1 upregulation in CRC cell lines. In the xenograft animal model, increased expression of TUG1 stimulated colony formation, migratory ability, and metastatic potential. Indeed, the researchers observed that TUG1 activated EMT-correlated gene expression [24, 77]. Another study proposed that the highly-expression of TUG1 was a CRC convinced unfavorable prognosis marker [78]. Cumulatively, TUG1 may act as a prognostic biomarker and a curative target. With more attempts affirm to the study of lncRNA particularly TUG1, it is optimistic that TUG1 will finally attain clinical utility [79]. In contrast, a recent study on 47 CRC patients indicated that there were no remarkable correlations between TUG1 expression and clinicopathological features of CRC. Besides, TUG1 expression could not forecast the overall survival and progression-free survival in CRC patients [80].
Conclusion
LncNAs can be used as biomarkers for the diagnosis, prognosis, and monitoring of the progression of the disease because of their tissue-specific expression and high stability [81]. Several studies reported that lncRNAs are closely linked to a variety of cancer types and might function as oncogenes or oncogene suppressors depending on the type of cancer [82, 83]. Although the role of TUG1 in the characteristics and chemoresistance of CRC stem cells is still not well-defined, it has been already presented as an attractive potential biomarker because of its tumor-promotive function via diverse mechanisms, such as RNA-RNA and RNA/transcription factors interactions [46, 84]. It was investigated by the same group that TUG1 increased the characteristics and oxaliplatin resistance of CRC stem cells by enhancing GATA6 stability [14]. TUG1 is suggested to solve the problem of fluoropyrimidine (Fu)-based chemotherapy. TUG1 appears to mediate 5-Fu resistance in CRC through the miR-197-3p/TYMS axis [9]. Knockdown of TUG1 resensitized resistant cells to the exposure of 5-Fu and induced cell apoptosis. This lncRNA by targeting miR-186 stimulated CPEB2 to mediate methotrexate resistance in CRC [15]. Taken together, the role of lncRNA TUG1 in CRC drug resistance seems to be crucial and holds great promise as a potential therapeutic target. Current findings regarding TUG1 not only promote a better understanding of CRC pathogenesis and development but also facilitated the progress of cancer lncRNA therapy. However, many mechanisms remain poorly described suggesting a great need for further study.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Sharma A, Yadav D, Rao P, Sinha S, Goswami D, Rawal RM, Shrivastava N. Identification of potential therapeutic targets associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Computers in Biology and Medicine 2022:105688.
Ottaiano A, Santorsola M, Perri F, Pace U, Marra B, Correra M, Sabbatino F, Cascella M, Petrillo N, Ianniello M. Clinical and molecular characteristics of rare malignant tumors of colon and rectum. Biology. 2022;11:267.
Wang L, Zhao Z, Feng W, Ye Z, Dai W, Zhang C, Peng J, Wu K. Long non-coding RNA TUG1 promotes colorectal cancer metastasis via EMT pathway. Oncotarget. 2016;7:51713.
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. Cancer J Clin. 2020;70:145–64.
Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Przeglad gastroenterologiczny. 2019;14:89–103.
Guo M, Peng Y, Gao A, Du C, Herman JG. Epigenetic heterogeneity in cancer. Biomark Res. 2019;7:23.
Testa U, Pelosi E, Castelli G: Colorectal cancer: genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Medical sciences (Basel, Switzerland) 2018, 6:31.
Zhunussova G, Afonin G, Abdikerim S, Jumanov A, Perfilyeva A, Kaidarova D, Djansugurova L. Mutation Spectrum of Cancer-Associated Genes in Patients With Early Onset of Colorectal Cancer. Frontiers in Oncology 2019, 9.
Song M, Chan AT. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clinical gastroenterology and hepatology: the official clinical practice. J Am Gastroenterological Association. 2019;17:275–89.
Xie Y-H, Chen Y-X, Fang J-Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Therapy. 2020;5:22.
Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:197.
Folprecht G, Martinelli E, Mazard T, Modest DP, Tsuji A, Esser R, Cremolini C, Falcone A. Triplet chemotherapy in combination with anti-EGFR agents for the treatment of metastatic colorectal cancer: Current evidence, advances, and future perspectives. Cancer Treat Rev. 2022;102:102301.
Florescu-Ţenea RM, Kamal AM, Mitruţ P, Mitruţ R, Ilie DS, Nicolaescu AC, Mogoantă L. Colorectal Cancer: An Update on Treatment Options and Future Perspectives. Curr health Sci J. 2019;45:134–41.
Herbst C-l, Miot JK, Moch SL, Ruff P. Colorectal Cancer (CRC) treatment and associated costs in the public sector compared to the private sector in Johannesburg, South Africa. BMC Health Serv Res. 2020;20:1–11.
Nam J, Kim S-W. MicroRNA as a Versatile Regulator of Wnt the Signaling Pathway in Colorectal Cancer. In: Biotechnologies for Gene Therapy. Springer; 2022. pp. 25–43.
Xu W-W, Jin J, Wu X-y, Ren Q-L, Farzaneh M. MALAT1-related signaling pathways in colorectal cancer. Cancer Cell Int. 2022;22:1–9.
Azizidoost S, Ghaedrahmati F, Anbiyaee O, Ahmad Ali R, Cheraghzadeh M, Farzaneh M. Emerging roles for lncRNA-NEAT1 in colorectal cancer. Cancer Cell Int. 2022;22:1–10.
Anbiyaiee A, Ramazii M, Bajestani SS, Meybodi SM, Keivan M, Khoshnam SE, Farzaneh M. The function of LncRNA-ATB in cancer. Clinical and Translational Oncology 2022:1–9.
Cao Z, Oyang L, Luo X, Xia L, Hu J, Lin J, Tan S, Tang Y, Zhou Y, Cao D. The roles of long non-coding RNAs in lung cancer. J Cancer. 2022;13:174.
Yin X, Lin H, Lin L, Miao L, He J, Zhuo Z. LncRNAs and CircRNAs in cancer. MedComm. 2022;3:e141.
Xu Y, Wang J, Qiu M, Xu L, Li M, Jiang F, Yin R, Xu L. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumor Biology. 2015;36:1643–51.
Zhang E, He X, Yin D, Han L, Qiu M, Xu T, Xia R, Xu L, Yin R, De W. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109–9.
Huang M-D, Chen W-M, Qi F-Z, Sun M, Xu T-P, Ma P, Shu Y. -q: Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14:1–12.
Sun J, Ding C, Yang Z, Liu T, Zhang X, Zhao C, Wang J. The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition. J translational Med. 2016;14:1–10.
Duan W, Nian L, Qiao J, Liu N. LncRNA TUG1 aggravates the progression of cervical cancer by binding PUM2. Eur Rev Med Pharmacol Sci. 2019;23:8211–8.
Karali M, Banfi S. Non-coding RNAs in retinal development and function. Hum Genet. 2019;138:957–71.
Lai L, Wang Y, Peng S, Guo W, Li F, Xu S. P53 and taurine upregulated gene 1 promotes the repair of the DeoxyriboNucleic Acid damage induced by bupivacaine in murine primary sensory neurons. Bioengineered. 2022;13:7439–56.
Zhou H, Sun L, Wan F. Molecular mechanisms of TUG1 in the proliferation, apoptosis, migration and invasion of cancer cells (Review). Oncol Lett. 2019;18:4393–402.
Miller SA, Damle M, Kingston RE. H3K27me3 is dispensable for early differentiation but required to maintain differentiated cell identity. bioRxiv 2020:2020.2006.2027.175612.
Zhou H, Sun L, Wan F. Molecular mechanisms of TUG1 in the proliferation, apoptosis, migration and invasion of cancer cells. Oncol Lett. 2019;18:4393–402.
Bencivenga D, Stampone E, Vastante A, Barahmeh M, Della Ragione F, Borriello A. An Unanticipated Modulation of Cyclin-Dependent Kinase Inhibitors: The Role of Long Non-Coding RNAs. Cells. 2022;11:1346.
Zilio N, Codlin S, Vashisht AA, Bitton DA, Head SR, Wohlschlegel JA, Bähler J, Boddy MN. A novel histone deacetylase complex in the control of transcription and genome stability. Mol Cell Biol. 2014;34:3500–14.
Wang H, Liao S, Li H, Chen Y, Yu J. Long Non-coding RNA TUG1 Sponges Mir-145a-5p to Regulate Microglial Polarization After Oxygen-Glucose Deprivation. Front Mol Neurosci. 2019;12:215–5.
Wang X, Chen X, Zhang D, Yang G, Yang Z, Yin Z, Zhao S. Prognostic and clinicopathological role of long non-coding RNA taurine upregulated 1 in various human malignancies: a systemic review and meta-analysis. Tumor Biology. 2017;39:1010428317714361.
Yang B, Tang X, Wang Z, Sun D, Wei X, Ding Y. TUG1 promotes prostate cancer progression by acting as a ceRNA of miR-26a. Biosci Rep 2018, 38.
Xie C, Chen B, Wu B, Guo J, Cao Y. LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in osteosarcoma by regulating miR-212-3p/FOXA1 axis. Biomed Pharmacother. 2018;97:1645–53.
Zhou Y, Lu Y, Li R, Yan N, Li X, Dai T. Prognostic role of long non-coding RNA TUG1 expression in various cancers: a meta-analysis. Oncotarget. 2017;8:100499.
Tang Q, Li X, Chen Y, Long S, Yu Y, Sheng H, Wang S, Han L, Wu W. Solamargine inhibits the growth of hepatocellular carcinoma and enhances the anticancer effect of sorafenib by regulating HOTTIP-TUG1/miR‐4726‐5p/MUC1 pathway. Mol Carcinog. 2022;61:417–32.
Tan J, Qiu K, Li M, Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015;589:3175–81.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90.
Zhai H-y, Sui M-h, Yu X, Qu Z, Hu J-c, Sun H-q, Zheng H-t, Zhou K. Jiang L-x: Overexpression of long non-coding RNA TUG1 promotes colon cancer progression. Med Sci monitor: Int Med J experimental Clin Res. 2016;22:3281.
Wang X, Bai X, Yan Z, Guo X, Zhang Y. The lncRNA TUG1 promotes cell growth and migration via the TUG1/miR-145-5p/TRPC6 pathway in colorectal cancer. 2020.
Yan Z, Bi M, Zhang Q, Song Y, Hong S. LncRNA TUG1 promotes the progression of colorectal cancer via the miR-138-5p/ZEB2 axis. Bioscience Reports 2020, 40.
Sun J, Ding C, Yang Z, Liu T, Zhang X, Zhao C, Wang J. Correction to: The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition. J Translational Med. 2020;18:1–6.
Li M-Z, Wang J-J, Yang S-B, Li W-F, Xiao L-B, He Y-L, Song X-M. ZEB2 promotes tumor metastasis and correlates with poor prognosis of human colorectal cancer. Am J translational Res. 2017;9:2838.
Xiao CH, Yu HZ, Guo CY, Wu ZM, Cao HY, Li WB, Yuan JF. Long non–coding RNA TUG1 promotes the proliferation of colorectal cancer cells through regulating Wnt/β–catenin pathway. Oncol Lett. 2018;16:5317–24.
Bian J, Dannappel M, Wan C, Firestein R. Transcriptional regulation of Wnt/β-catenin pathway in colorectal cancer. Cells. 2020;9:2125.
Itatani Y, Kawada K, Sakai Y. Transforming growth factor-β signaling pathway in colorectal cancer and its tumor microenvironment. Int J Mol Sci. 2019;20:5822.
Shen X, Hu X, Mao J, Wu Y, Liu H, Shen J, Yu J, Chen W. The long noncoding RNA TUG1 is required for TGF-β/TWIST1/EMT-mediated metastasis in colorectal cancer cells. Cell Death Dis. 2020;11:1–10.
Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28.
Liu W, Meng J, Su R, Shen C, Zhang S, Zhao Y, Liu W, Du J, Zhu S, Li P. SP1-mediated up-regulation of lncRNA TUG1 underlines an oncogenic property in colorectal cancer. Cell Death Dis. 2022;13:1–10.
Bajpai R, Nagaraju GP. Specificity protein 1: Its role in colorectal cancer progression and metastasis. Crit Rev Oncol/Hematol. 2017;113:1–7.
Li G, Song H, Chen L, Yang W, Nan K, Lu P. TUG1 promotes lens epithelial cell apoptosis by regulating miR-421/caspase-3 axis in age-related cataract. Exp Cell Res. 2017;356:20–7.
Xue L, Yang D. MiR-421 inhibited proliferation and metastasis of colorectal cancer by targeting MTA1. J BUON. 2018;23:1633–9.
Cao L-L, Du C, Liu H, Pei L, Qin L, Jia M, Wang H. Lysine-specific demethylase 2A expression is associated with cell growth and cyclin D1 expression in colorectal adenocarcinoma. Int J Biol Mark. 2018;33:407–14.
Byrne DP, Foulkes DM, Eyers PA. Pseudokinases: update on their functions and evaluation as new drug targets. Future Med Chem. 2017;9:245–65.
Hou Z, Guo K, Sun X, Hu F, Chen Q, Luo X, Wang G, Hu J, Sun L. TRIB2 functions as novel oncogene in colorectal cancer by blocking cellular senescence through AP4/p21 signaling. Mol Cancer. 2018;17:1–15.
Liu Q, Zhang W, Luo L, Han K, Liu R, Wei S, Guo X. Long noncoding RNA TUG1 regulates the progression of colorectal cancer through miR-542-3p/TRIB2 axis and Wnt/β-catenin pathway. Diagn Pathol. 2021;16:1–12.
Zhu G-X, Gao D, Shao Z-Z, Chen L, Ding W-J, Yu Q-F. Wnt/β–catenin signaling: Causes and treatment targets of drug resistance in colorectal cancer. Mol Med Rep. 2021;23:1–1.
Shao H, Dong D, Shao F. Long non-coding RNA TUG1-mediated down-regulation of KLF4 contributes to metastasis and the epithelial-to-mesenchymal transition of colorectal cancer by miR-153-1. Cancer Manage Res. 2019;11:8699.
Ma Y, Wu L, Liu X, Xu Y, Shi W, Liang Y, Yao L, Zheng J, Zhang J. KLF4 inhibits colorectal cancer cell proliferation dependent on NDRG2 signaling. Oncol Rep. 2017;38:975–84.
Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–87.
Zhu Y, Hu H, Yuan Z, Zhang Q, Xiong H, Hu Z, Wu H, Huang R, Wang G, Tang Q. LncRNA NEAT1 remodels chromatin to promote the 5-Fu resistance by maintaining colorectal cancer stemness. Cell Death Dis. 2020;11:1–11.
Joag MG, Sise A, Murillo JC, Sayed-Ahmed IO, Wong JR, Mercado C, Galor A, Karp CL. Topical 5-fluorouracil 1% as primary treatment for ocular surface squamous neoplasia. Ophthalmology. 2016;123:1442–8.
Wang M, Hu H, Wang Y, Huang Q, Huang R, Chen Y, Ma T, Qiao T, Zhang Q, Wu H. Long non-coding RNA TUG1 mediates 5-fluorouracil resistance by acting as a ceRNA of miR-197-3p in colorectal cancer. J Cancer. 2019;10:4603.
Tarazona N, Gimeno-Valiente F, Gambardella V, Huerta M, Roselló S, Zuniga S, Calon A, Carbonell-Asins JA, Fontana E, Martinez-Ciarpaglini C. Detection of postoperative plasma circulating tumour DNA and lack of CDX2 expression as markers of recurrence in patients with localised colon cancer. ESMO open. 2020;5:e000847.
Sun J, Zhou H, Bao X, Wu Y, Jia H, Zhao H, Liu G: lncRNA TUG1 Facilitates Colorectal Cancer Stem Cell Characteristics and Chemoresistance by Enhancing GATA6 Protein Stability. Stem Cells International 2021, 2021.
Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG, Erdkamp FL, Vos AH, van Groeningen CJ, Sinnige HA. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med. 2009;360:563–72.
Li C, Gao Y, Li Y, Ding D. TUG1 mediates methotrexate resistance in colorectal cancer via miR-186/CPEB2 axis. Biochem Biophys Res Commun. 2017;491:552–7.
Huang X, Zhang H, Guo X, Zhu Z, Cai H, Kong X. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J Hematol Oncol. 2018;11:1–15.
Zhi S, Li J, Kong X, Xie X, Zhang Q, Fang G. Insulin-like growth factor 2 mRNA binding protein 2 regulates proliferation, migration, and angiogenesis of keratinocytes by modulating heparanase stability. Bioengineered. 2021;12:11267–76.
Fakhraldeen SA, Clark RJ, Roopra A, Chin EN, Huang W, Castorino J, Wisinski KB, Kim T, Spiegelman VS, Alexander CM. Two isoforms of the RNA binding protein, coding region determinant-binding protein (CRD-BP/IGF2BP1), are expressed in breast epithelium and support clonogenic growth of breast tumor cells. J Biol Chem. 2015;290:13386–400.
Xia C, Li Q, Cheng X, Wu T, Gao P, Gu Y. Insulin-like growth factor 2 mRNA-binding protein 2-stabilized long non-coding RNA Taurine up-regulated gene 1 (TUG1) promotes cisplatin-resistance of colorectal cancer via modulating autophagy. Bioengineered. 2022;13:2450–69.
Yang Y, Yan X, Li X, Ma Y, Goel A. Long non-coding RNAs in colorectal cancer: Novel oncogenic mechanisms and promising clinical applications. Cancer Lett. 2021;504:67–80.
Talebi A, Azizpour M, Agah S, Masoodi M, Mobini GR, Akbari A. The relevance of long noncoding RNAs in colorectal cancer biology and clinical settings. J Cancer Res Ther. 2020;16:22–3.
Li N, Shi K, Kang X, Li W. Prognostic value of long non-coding RNA TUG1 in various tumors. Oncotarget. 2017;8:65659.
Saus E, Brunet-Vega A, Iraola-Guzman S, Pegueroles C, Gabaldon T, Pericay C. Long non-coding RNAs as potential novel prognostic biomarkers in colorectal cancer. Front Genet. 2016;7:54.
Liu J, Lin J, Li Y, Zhang Y, Chen X. Prognostic role of lncRNA TUG1 for cancer outcome: evidence from 840 cancer patients. Oncotarget. 2017;8:50051.
Li Z, Shen J, Chan MT, Wu WKK. TUG 1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49:471–5.
Abushouk AI, Kattan SW, Ahmedah HT, Baothman E, Shaheen S, Toraih EA, Fawzy MS. Expression of oncolong noncoding RNA taurine-upregulated gene-1 in colon cancer: A clinical study supported by in silico analysis. J Can Res Ther 2021.
Kunej T, Obsteter J, Pogacar Z, Horvat S, Calin GA. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 2014;51:344–57.
Wei S, Sun S, Zhou X, Zhang C, Li X, Dai S, Wang Y, Zhao L, Shan B. SNHG5 inhibits the progression of EMT through the ubiquitin-degradation of MTA2 in oesophageal cancer. Carcinogenesis. 2021;42:315–26.
Gong M, Wang X, Mu L, Wang Y, Pan J, Yuan X, Zhou H, Xing J, Wang R, Sun J. Steroid receptor coactivator-1 enhances the stemness of glioblastoma by activating long noncoding RNA XIST/miR‐152/KLF4 pathway. Cancer Sci. 2021;112:604–18.
Sun J, Zhou H, Bao X, Wu Y, Jia H, Zhao H, Liu G: lncRNA TUG1 Facilitates Colorectal Cancer Stem Cell Characteristics and Chemoresistance by Enhancing GATA6 Protein Stability. Stem cells international 2021, 2021:1075481–1075481.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
SH. A., A. N., F. GH., K. R., B. K., P. M., and M. F., have made contributions to the writing of the manuscript. All authors have approved the submitted version of the article and have agreed to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there is no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Azizidoost, S., Nasrolahi, A., Ghaedrahmati, F. et al. The pathogenic roles of lncRNA-Taurine upregulated 1 (TUG1) in colorectal cancer. Cancer Cell Int 22, 335 (2022). https://doi.org/10.1186/s12935-022-02745-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-022-02745-1