Abstract
Background
Efforts to construct the Streptomyces host strain with enhanced yields of heterologous product have focussed mostly on engineering of primary metabolism and/or the deletion of endogenous biosynthetic gene clusters. However, other factors, such as chromosome compactization, have been shown to have a significant influence on gene expression levels in bacteria and fungi. The expression of genes and biosynthetic gene clusters may vary significantly depending on their location within the chromosome. Little is known about the position effect in actinomycetes, which are important producers of various industrially relevant bioactive molecules.
Results
To demonstrate an impact of the chromosomal position effect on the heterologous expression of genes and gene clusters in Streptomyces albus J1074, a transposon mutant library with randomly distributed transposon that includes a β-glucuronidase reporter gene was generated. Reporter gene expression levels have been shown to depend on the position on the chromosome. Using a combination of the transposon system and a φC31-based vector, the aranciamycin biosynthetic cluster was introduced randomly into the S. albus genome. The production levels of aranciamycin varied up to eightfold depending on the location of the gene cluster within the chromosome of S. albus J1074. One of the isolated mutant strains with an artificially introduced attachment site produced approximately 50% more aranciamycin than strains with endogenous attBs.
Conclusions
In this study, we demonstrate that expression of the reporter gene and aranciamycin biosynthetic cluster in Streptomyces albus J1074 varies up to eightfold depending on its position on the chromosome. The integration of the heterologous cluster into different locations on the chromosome may significantly influence the titre of the produced substance. This knowledge can be used for the more efficient engineering of Actinobacteria via the relocation of the biosynthetic gene clusters and insertion of additional copies of heterologous constructs in a suitable chromosomal position.
Similar content being viewed by others
Background
During the past few decades, streptomycetes have been extensively employed for the heterologous expression of bioactive natural products. Heterologous expression contributes substantially to the discovery and characterisation of secondary metabolites. The increase in the number of sequenced microbial genomes demonstrates that most of the biosynthetic clusters remain silent under standard laboratory conditions. The expression of silent gene clusters in a well-characterised heterologous host is a validated approach for the discovery of new natural products [1]. However, insufficient yields of heterologously produced molecules often prevent their further purification and characterisation. Currently, several Streptomyces strains have been utilised as hosts for the production of natural products. The most widely used are Streptomyces coelicolor, Streptomyces avermitillis and Streptomyces albus J1074 [2]. However, the production level of the heterologously produced compounds often needs to be further optimised. Industrially relevant titres of some natural products may reach the multi-gram per litre scale (e.g., ≥10 g/L erythromycin; ≥30 g/L tetracycline; ≥80 g/L salinomycin; personal communication with Prof. Hrvoje Petkovic, Ljubljana University), whereas the yields of natural products in heterologous hosts remain in the milligram range [2]. One of the factors significantly influencing the expression of genes and biosynthetic gene clusters is their location on the chromosome, the so-called ‘chromosomal position effect’. This term describes differences in the expression of a gene depending on its chromosomal location. Studies of gene expression in Saccharomyces cerevisiae demonstrated that the expression of heterologous genes was repressed when integrated at silent loci, indicating that repression occurred due to a positional effect [3]. In another study [4], it was shown that an Aspergillus nidulans mutant, defective in the system responsible for histone methylation, activates silent secondary metabolite clusters.
The position effect can influence not only the expression level of a native gene after spontaneous translocations, but also transgene expression after insertion into different regions of a genome, leading to changes in heterologous production. It was demonstrated that variations in β-galactosidase activity can reach 300-fold in response to translocations of the β-galactosidase gene in the Escherichia coli chromosome [5]. However, Schmid and Roth demonstrated [6] only a threefold variation in the expression level of the his operon cluster randomly distributed within the Salmonella typhimurium chromosome. Additionally, Lactobacillus lactis mutants showed a threefold difference in the levels of gus(a) expression [7].
The main factors causing such variability in gene expression are: (i) the level of DNA compactization [8]; (ii) variations in the promoter strength; and (iii) the distance to the origin of replication [7]. The first two factors are more critical for eukaryotic organisms, which have high levels of DNA compactization and greater variation in promoter strengths, which may influence a downstream heterologous gene. By contrast, the distance to the replication origin is the major factor of variability in gene expression in prokaryotic cells, because they contain a single origin of replication per genophore. Thus, a gene placed closer to the origin of replication is replicated before a gene located near the terminus and therefore has an operative increase in gene dosage [9].
Despite the importance of streptomycetes as producers of natural products, the position effect has not yet been investigated in them. The representatives of this genus have a complex life cycle. These bacteria colonise the environment by growing branching, multigenomic hyphae while simultaneously forming unigenomic spores to achieve dispersion. Recent studies identified a novel protein specific to Actinobacteria that is responsible for DNA compactization, chromosome segregation and antibiotic production [10]. In other studies [11], a histone-like protein was characterised that can structurally couple changes in DNA conformation and transcription in response to stress. Such complexity in nucleoid organisation leads to variations in expression of different DNA regions and causes a chromosomal position effect. However, to date, a lack of efficient tools for the exploration of streptomycetes genomics has limited any attempts to evaluate the impact of this factor on the expression of heterologous genes and clusters.
Here, we extend previous studies on the chromosomal position effect to the mycelial microorganism Streptomyces albus J1074, which is a typical representative of the Streptomyces genus and is well known for its outstanding potential as a host for the heterologous production of bioactive small molecules [12]. During the past decade, S. albus J1074 has been used for the heterologous expression of various antibiotic biosynthetic clusters, e.g. thiocoraline, cyclooctatin and steffimycin. Numerous genetic tools have recently been developed for the strain, making its genome engineering rather straightforward [13–15].
To evaluate the impact of the chromosomal position effect on heterologous expression in the genome of S. albus J1074, a transposon mutant library with randomly distributed gus(a), the β-glucuronidase reporter gene, was generated. The expression of the reporter gene in recombinant strains was analysed, and various factors influencing the expression of the gusA gene are discussed. In addition, using a combination of a transposon system and the φC31-based vector, the aranciamycin biosynthetic cluster was introduced randomly into the S. albus genome. Analysis of aranciamycin producers also demonstrated an eightfold variation in the production level of the antibiotic depending on its chromosomal location. One of the isolated mutant strains with an artificially introduced attachment site produced 53% more aranciamycin than strains with endogenous attBs.
Results and discussion
Impact of the chromosomal position effect on gene expression
To examine an impact of the chromosomal position effect on the expression of heterologous genes in S. albus J1074, a transposon harbouring the reporter gene was constructed (Fig. 1). The transposon contained the gus(a) reporter gene, encoding β-glucuronidase under the control of the ermEp1 promoter [16]. The activity of β-glucuronidase can be spectrophotometrically measured. The gus(a) gene is flanked by two fd-phage terminators [17], which prevent transcription in both directions with an efficiency of up to 85% (Additional file 1: Figure S1). The apramycin resistance gene was cloned upstream and in the opposite direction to gus(a) to exclude any impact on expression of the reporter gene. Then the transposon was cloned into the plasmid containing synthetic gene of Himar1-transposase and temperature sensitive pSG5 replicon. The vector was introduced into genome S. albus J1074 and library of transposon mutants was generated as described previously [14]. Eighteen mutants from this library with the randomly distributed reporter gene were isolated and analysed. The insertions were mapped on the chromosome using rescue cloning, as described elsewhere [14] (Table 1; Fig. 2). All identified insertions were localised in a core region of the chromosome. A measurement of the GusA activities was made after 48 h of growth (Table 1; Fig. 4). The average value of activity was 8.6 U/mg, and eight of the analysed mutants (44%) had activities that lay within this range (mutants # 1, 2, 4, 5, 14, 16, 17 and 18). Five mutants (28%) had activity values above average, and another five mutants (28%), located mostly in the centre of the chromosome, and had values below average. The variation in the reporter gene expression was six-fold (the lowest value observed was 2.7 U/mg, and the highest was 17.3 U/mg), which was comparable to results described for other prokaryotes, where two- to threefold variations in activity have been observed [6, 7, 18].
The map of the plasmid containing gus(a) in the transposon, pALG. Plasmids contains following features: himar1(a)—synthetic transposase gene, under control of p1—tipAp, thiostrepton inducible promoter; gus(a)—reporter gene of glucuronidase; p2—ermEp1, promoter 1 of erythromycin resistance gene; tfd terminator of fd-phage, pSG5rep actinomycetes temperature-sensitive replicon; aac(3)IV apramycin resistance marker, hph hygromycin resistance marker, ITR inverted terminal repeats; R6Kori origin for rescue cloning
Owing to the isolation of gus(a) by the two terminators, the activity of the local promoters should not have had a significant impact on its expression. To confirm this, the GusA activities of the mutants were compared with S. albus J1074 RNA-seq data after 48 h of cultivation in liquid medium. The RPKM (reads per kilobase per million reads) values of the genes, which are located in the genome of S. albus J1074 upstream and in the same orientation as gus(a), were analysed. The results demonstrated that, in the obtained transposon mutants, local promoters had a minor effect on the level of gus(a) expression, e.g., mutant 10, which exhibited one of the lowest levels of GusA activity, was flanked upstream by the region with a high RPKM value (Fig. 3). Concurrently, mutants with a relatively low RPKM of flanking genes (e.g. mutants 7 and 15) were characterised by a high GusA activity level (Fig. 3).
The comparison of GusA-activity levels with expression level of adjacent genes (correlation coefficient is equal—0.34). Blue columns correspond to values of GusA-activity. Strains were grown for 48 h at 28 °C; light grey columns correspond to RPKM after 48 h of cultivation at 28 °C. Mutants are placed according to location of their transposons on the chromosome
Chromosomal position effect does not correlate with distance to oriC
Previous studies on the position effect in E. coli and Salmonella demonstrated that variations in the expression level of reporter genes are caused mainly by gene dosage [6, 19–21]. Unlike most bacteria that possess a circular chromosome, streptomycetes carry a linear chromosome with a centrally located oriC. Thus, if the gene dosage plays a significant role in the expression level of heterologous genes, the GusA activity in mutants with insertions localised closer to the chromosome centre should be higher than in those with insertions localised closer to the chromosome ends. However, plotting the GusA activities with positions of the reporter gene showed that mutants with integration near the oriC demonstrated GusA activity below average (mutants 8, 9, 10, 11 and 12), whereas strains with an insertion located some distance from the oriC, closer to centres of the chromosomal shoulders, demonstrated above-average results (mutants 3, 6 and 15).
Relocation of the site for gene cluster integration
The obtained results demonstrated that the translocation of the heterologous construct may significantly influence its expression. This result should also be valid for more complex genetic structures, such as an antibiotic biosynthetic gene cluster. It is difficult to deliver antibiotic biosynthetic gene clusters into a streptomycetes chromosome via a transposon, due to their large size. Most of the vectors carrying biosynthetic gene clusters are integrated into the chromosome via the phage φC31 integration system. Thus, we translocated the attB attachment site of φC31 into various locations of the S. albus J1074 chromosome. A transposon with an attachment site of the φC31-phage was constructed and cloned into a non-replicative vector containing himar1(a), the synthetic transposase gene. The obtained plasmid pAHT (Fig. 4) did not require any additional induction steps or curing of the plasmid backbone, because the exconjugants obtained already contained a copy of the transposon inserted in the genome. This vector was than introduced into S. albus SAM3 (ΔattB·ΔpseB4), lacking both endogenous attB sites [22], by intergeneric conjugation from E. coli.
The map of the plasmid containing phage attachment site in the transposon, pAHT. Plasmids contains following features: himar1(a)—synthetic transposase gene, under control of p1—tipAp, thiostrepton inducible promoter, aac(3)IV apramycin resistance marker, hph hygromycin resistance marker, ITR inverted terminal repeats, R6Kori origin for rescue cloning, attB attachment site of φC31-phage
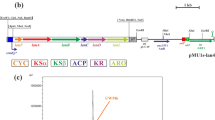
The aranciamycin gene cluster was integrated into artificially distributed attBs. This 35.9-kb large cluster was isolated from Streptomyces echinatus [23] and was previously expressed in different streptomycetes [24]. The cosmid with the aranciamycin cluster was introduced into twenty-four transposon mutants. In this way, a library of strains carrying the aranciamycin biosynthetic cluster at different chromosomal positions was obtained. As a control, two strains containing naturally located attBs (ΔattB and ΔpseB4 [22]) were used. Altogether, the mutants demonstrated an eightfold variation in aranciamycin production. Such a variation in the expression level of a heterologous cluster correlated with results obtained for gus(a)-gene expression described herein. The average production of aranciamycin for the mutant strains was 40.53 U/mg. However, in this case, we observed stronger variation in the results: only one of the 24 mutants produced in the range of 40.53 ± 10% (mutant 24). Thirteen mutants (54%) demonstrated below-average results, and 10 mutants (42%) evinced above-average results (Fig. 5). The production of aranciamycin from four artificially integrated attBs was more than 20% above the average production from endogenous attachment sites (Fig. 5, mutants 1, 2, 4 and 21). The yield of aranciamycin in one of those mutants (mutant 2) was 56% higher than in the control strains. This result demonstrated that the translocation of the cluster may result in significant variation in antibiotic production.
Relative concentration of aranciamycin in culture media of heterologous aranciamycin producers. dattB—S. albus SAM1(ΔattB)::p412C06; dpseB4—S. albus SAM2(ΔpseB4)::-p412C06; 1-24—S. albus SAM3 (ΔattB·ΔpseB4)::pAHT::p412C06-mutants. Dotted black line corresponds to average production of aranciamycin by transposon mutant strains; dotted red line corresponds to average production of aranciamycin by control strains with one integration site. Strains were grown for 120 h at 28 °C. Relative concentration of aranciamycin was calculated as peak area per gram of dry mass
These data suggest that the central regions of the chromosomal “shoulders” are more preferable loci for the expression of heterologous constructs, because GusA activity in mutants with these integrations was higher than average. Given that streptomycetes are well-established cell factories for the biosynthesis of natural products, it is important to study the influence of chromosomal localisation on the expression of biosynthetic gene clusters.
Indeed, the eightfold variation in antibiotic production was observed in the library of mutants carrying corresponding gene clusters in different locations of the chromosome. Four of the 24 isolated mutants demonstrated more than a 20% increase in antibiotic production, and one of these mutants showed a 56.5% increase in the yield of aranciamycin (82.4 U/mg) compared with the average production of the control strains (52.65 U/mg).
Thus, the proposed approach can be used in combination with other methods that are employed for the optimisation of the heterologous production of natural products. This approach can be used not only for translocation of the attachment site, but also to introduce additional integration sites into the genome, thus resulting in an increase in the number of clusters per genome [25]. We believe that the genetic tools described herein will find wide applications in the construction and manipulation of actinobacterial heterologous hosts.
Conclusions
Our study demonstrates the effect of the chromosomal position effect on a heterologously expressed reporter gene and on more complex genetic constructs, such as antibiotic biosynthetic gene clusters in S. albus J1074. Based on this phenomenon, we developed an approach that can be used for the construction of host strains with improved heterologous production features.
Methods
Bacterial strains and cultivation conditions
The cultivation of the S. albus strains was conducted on a soy-mannitol agar or in a liquid TSB medium at 29 °C. The cultivation of E. coli was conducted in an LB medium at 37 °C. The intergeneric conjugation of the plasmids was performed as previously described [26]. The library of transposon mutants was obtained as described elsewhere [14].
Recombinant DNA techniques
Lambda-red-mediated recombination was conducted as described in the standard protocol [27]. The isolation of genomic DNA from S. albus strains was performed according to a previously described method [26]. PCR using Phusion polymerase (Thermo Scientific), separation of DNA by agarose gel electrophoresis and Southern blot hybridisation were performed as previously described [28]. DNA fragments and PCR products were purified using PROMEGA™ kits. Oligonucleotides were synthesised by Eurofins MWG Operon. DNA sequencing was performed by GATC-Biotec.
Isolation of rescue plasmids
Chromosomal DNA was isolated from the desired strains and digested with SacII for pALG transconjugants, respectively. The fragments were then self-ligated and transformed into E. coli Transformax cells. The transformants were selected based on their resistance to apramycin. The obtained plasmids were isolated and sequenced using the P1-pALG-ch primer.
Plasmid construction
The plasmids and primers used in this study are listed below (see Additional file 1). The DNA manipulations and cloning procedures were performed according to standard protocols [29], and the plasmid constructs were confirmed by DNA sequencing.
Construction of pALG
The gus(a) gene was amplified using the pSETgus(a) plasmid as a template, the forward primer, Fr-XI-ep1-gus(a), carrying the XbaI site and ermEp1, a Saccharopolyspora erythraea ermEp1 gene promoter and the reverse primer, Rs-MI-tfd-gus(a), carrying the fd-phage terminator and the MunI site. The PCR fragment was ligated into pNheIaac [14] linearised by XbaI and MunI leading to p11-8. The aac(3)IV gene was amplified using pIJ773 as a template, the forward primer, Fr-ERI-tfd-aac, carrying the fd terminator and the reverse primer, Rs-ERI-aac. Both primers carry the EcoRI site. The fragment was cloned into the EcoRI site of p11-8 replacing an existing aac(3)IV, resulting in p11-8aactfd. The PvuII fragment of p11-8aactfd, containing the transposon, was ligated to pALHim linearised with EcoRV, to yield pALG (Fig. 1).
Construction of pAHT
The hph gene was amplified using pAL1 as a template, with Fr-MI-attB-hph as the forward primer, carrying attB and the MunI restriction site, and Rs-XI-hph as the reverse primer carrying the XbaI site. The amplified fragment was cloned into the MunI and XbaI sites of pTn5Oks resulting in pTn5OksattBhph(II). To construct a backbone of the transposon vector, the hygromycin resistance gene in p31Him was replaced by the apramycin resistance gene using λ-red-mediated recombination [30]. The primers Fr-hph/aac(3)IV and Rs-hph/aac(3)IV were used to amplify the fragment for recombination. The obtained plasmid, pAHS, was linearized by EcoRV and ligated with the transposon from the pTn5OksattBhph(II) plasmid, giving pAHT (Fig. 4).
Measurement of glucuronidase activity
After a stationary phase of growth was reached, 10 mL of the main culture was transferred into 15 mL-conical centrifuge tubes. Mycelium was pelleted by centrifugation, the supernatant was discarded and the samples were dried for 24 h at 65 °C. After 24 h, the conical centrifuge tubes with dry mycelium were weighed. The spectrophotometric measurement of glucuronidase activity was carried out as previously described [29].
Strain cultivation and HPLC analysis
The S. albus strains containing the aranciamycin biosynthetic cluster were grown in 20 mL of a TSB medium for 48 h at 30 °C. Then, 200 μL of preculture were transferred into 15 mL of a NL5 + 1% yeast extract medium and cultivated for an additional 120 h at 30 °C. After cultivation, 5 mL of the mycelia were harvested by centrifugation for 30 min at 2000 rcf. The supernatant was transferred into a new Falcon tube and mixed with 5 mL of ethyl acetate and rotated on a rotator for 30 min. Then, the samples were centrifuged at 4000 rpm for 10 min, and the upper phase of the samples was transferred to glass vials and dried under N2. The dry pellet was dissolved in acetonitrile and centrifuged at 20,000 rcf for 10 min to remove the cell debris. Then, 80 μL of the extracts were transferred into HPLC vials and analysed by HPLC.
The HPLC–ESI–MS-UV–Vis analysis was performed on a Dionex Ultimate 3000 HPLC system (Thermo Fisher Scientific) that was connected to an ESI–MS Amazon mass spectrometer (Bruker). The HPLC system was equipped with a 100 × 2.1 mm, 1.7-µm BEH C18 column (Waters); a 50 × 2.1 mm, 1.7-µm BEH C18 column (Waters); or a 100 × 2 mm, 2.5-µm Luna C18 column (Phenomenex), depending on the method. For unknown extracts, an 18-min gradient was performed, mainly on the long column, whereas a 9-min gradient was used for the prepurified or known extracts. All gradient methods began with 5% B and increased to 95% B over 9 or 18 min. Solvent A consisted of H2O + 0.1% formic acid, whereas solvent B contained ACN+ 0.1% formic acid. The flow rate for the BEH C18 columns was 0.6 mL/min, whereas the flow rate for the Luna C18 column was 0.4 mL/min; UV–Vis detection was performed from 210 to 600 nm. Most of the time, the ESI–MS was used in alternating mode (positive and negative). Only a full MS scan or an additional scan with MS2 data was recorded, depending on the method. Both HPLC and MS systems were combined using the Hystar program (Bruker). As such, the standard LC–MS experiments could be selected as supermethods. These experiments included the selected column (Luna = Luna C18; 100 = BEH C18 100 mm; 50 = BEH C18 50 mm), gradient (9, 18 min) and MS mode (MS only, ms2 posneg).
High-resolution ESI–MS was measured using a Maxis Q-Tof 4G (Bruker) or an Orbitrap LTQ (Thermo Fisher Scientific) mass spectrometer connected to the same HPLC system used for standard LC–MS.
Abbreviations
- RPKM:
-
reads per kilobase of transcript per million reads
- Cons. hyp. prot.:
-
conservative hypothetical protein
- Reg. prot.:
-
regulatory protein
- IGR:
-
intergenic region
- Membr. prot.:
-
membrane protein
- ME:
-
mosaic end
- ITR:
-
inverted terminal repeat
References
Ongley SE, Bian X, Neilan BA, Müller R. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat Prod Rep. 2013;30:1121–38.
Baltz RH. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol. 2016;43:343–70.
Schnell R, Rine J. A position effect on the expression of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:494–501.
Bok JW, Chiang Y-M, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–4.
Bryant JA, Sellars LE, Busby SJW, Lee DJ. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014;. doi:10.1093/nar/gku828.
Schmid MB, Roth JR. Gene location affects expression level in Salmonella typhimurium. J Bacteriol. 1987;169:2872–5.
Thompson A, Gasson MJ. Location effects of a reporter gene on expression levels and on native protein synthesis in Lactococcus lactis and Saccharomyces cerevisiae. Appl Environ Microbiol. 2001;67:3434–9.
Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–62.
Pavitt GD, Higgins CF. Chromosomal domains of supercoiling in Salmonella typhimurium. Mol Microbiol. 1993;10:685–96.
Swiercz JP, Nanji T, Gloyd M, Guarne A, Elliot MA. A novel nucleoid-associated protein specific to the actinobacteria. Nucleic Acids Res. 2013;41:4171–84.
Aldridge M, Facey P, Francis L, Bayliss S, Del Sol R, Dyson P. A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress? Nucleic Acids Res. 2013;41:4813–24.
Baltz R. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol. 2010;37:759–72.
Petzke L, Luzhetskyy A. In vivo Tn5-based transposon mutagenesis of Streptomycetes. Appl Microbiol Biotechnol. 2009;83:979–86.
Bilyk B, Weber S, Myronovskyi M, Bilyk O, Petzke L, Luzhetskyy A. In vivo random mutagenesis of streptomycetes using mariner-based transposon Himar1. Appl Microbiol Biotechnol. 2013;97:351–9.
Siegl T, Luzhetskyy A. Actinomycetes genome engineering approaches. Antonie Van Leeuwenhoek. 2012;102:503–16.
Bibb MJ, White J, Ward JM, Janssen GR. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol Microbiol. 1994;14:533–45.
Otsuka J, Kunisawa T. Characteristic base sequence patterns of promoter and terminator sites in phi X174 and fd phage DNAs. J Theor Biol. 1982;97:415–36.
Sousa C, de Lorenzo V, Cebolla A. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology. 1997;143:2071–8.
Beckwith JR, Signer ER, Epstein W. Transposition of the lac region of E. coli. Cold Spring Harbor Symp Quant Biol. 1966;31:393–401.
Block DH, Hussein R, Liang LW, Lim HN. Regulatory consequences of gene translocation in bacteria. Nucleic Acids Res. 2012;40:8979–92.
Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli. J Mol Biol. 1968;31:519–40.
Bilyk B, Luzhetskyy A. Unusual site-specific DNA integration into the highly active pseudo-attB of the Streptomyces albus J1074 genome. Appl Microbiol Biotechnol. 2014;98:5095–104.
Keller-Schierlein W, Müller A. The sugar component of aranciamycin: 2-0-methyl-l-rhamnose. Experientia. 1970;26:929–30.
Luzhetskyy A, Mayer A, Hoffmann J, Pelzer S, Holzenkämper M, Schmitt B, Wohlert SE, Vente A, Bechthold A. Cloning and heterologous expression of the aranciamycin biosynthetic gene cluster revealed a new flexible glycosyltransferase. ChemBioChem. 2007;8:599–602.
Mandelscheid N, Bilyk B, Busche T, Kalinowski J, Paululat T, Bechthold A, Petzke L, Luzhetskyy A. An influence of the copy number of biosynthetic gene clusters on the production level of antibiotics in a heterologous host. J Biotechnol. 2016;20(232):110–7. doi:10.1016/j.jbiotec.2016.05.038.
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Strepromyces genetics. Norwich: John Innes Foundation; 2000.
Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–5.
Sambrook J, Russel DW. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001.
Myronovskyi M, Welle E, Fedorenko V, Luzhetskyy A. β-Glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol. 2011;77:5370–83.
Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–6.
Authors’ contributions
BB participated in the design of experiments, performed the experiments and analysed the data. LH performed additional verification of the results and analysed the data. AL designed the experiments and performed project supervision. BB and AL wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The constructed mutant strains and plasmids are available at the Department of Pharmaceutical Biotechnology of the Saarland University, Saarbrücken, Germany.
Funding
This work was supported through funding from DZIF grant and by the European Commission under the 7th Framework Program through the “Collaborative Project” action “STREPSYNTH” Grant No. 613877 and through the ERC starting grant EXPLOGEN No. 281623 to AL.
Author information
Authors and Affiliations
Corresponding author
Additional file
12934_2016_619_MOESM1_ESM.docx
Additional file 1: Figure S1. Glucuronidase activity in cell lysates of recombinant S. albus strains. Table S1. Plasmids used in this work. Table S2. Primers used in this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Bilyk, B., Horbal, L. & Luzhetskyy, A. Chromosomal position effect influences the heterologous expression of genes and biosynthetic gene clusters in Streptomyces albus J1074. Microb Cell Fact 16, 5 (2017). https://doi.org/10.1186/s12934-016-0619-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-016-0619-z