Abstract
Background
Increasing evidence revealed that high expression level of lncRNA SNHG1 was associated with the unfavorable prognosis of cancer and maybe used as a valuable biomarker for cancer patients. The present meta->analysis is to analyze existing data to reveal potential clinical application of SNHG1 on cancer prognosis and tumor progression. All of the included studies were collected through a variety of retrieval strategies. And the articles were qualified by MOOSE and PRISMA checklists.
Methods
Up to Mar 20, 2018, literature collection was performed by comprehensive search through electronic databases, including the Cochrane library, PubMed, Embase, Web of science, Springer, Science direct, and three Chinese databases: CNKI, Weipu, and Wanfang. We analyzed 14 studies that met the criteria, and concluded that the increased SHNG1 level was correlated with poor OS and tumor progression.
Results
The combined results indicated that elevated SNHG1 expression level was significantly associated with poor OS (HR = 2.06, 95% CI: 1.69–2.52, P < 0.01) and PFS (HR = 2.78, 95% CI: 1.69–4.55, P < 0.01) in various cancers. Moreover, the promoted SNHG1 expression was also associated with tumor progression ((III/IV vs. I/II: HR = 1.89, 95% CI: 1.53–2.34, P < 0.01). In stratified analyses, a significantly unfavorable association of elevated lncRNA SNHG1 and OS was observed in both digestive system (HR = 2.04, 95% CI: 1.56–2.68, P < 0.01) and non-digestive system (HR = 2.09, 95% CI: 1.55–2.83, P < 0.01) cancer patients.
Conclusions
The present analysis indicated that the increased SNHG1 is associated with poor OS in patients with general tumors and may be served as a useful prognostic biomarker.
Similar content being viewed by others
Background
Cancer has gradually becoming a major threaten for human health in the worldwide [1, 2]. Even though tremendous improvements had been made in cancer treatment, the long-term survival rate still remains unsatisfied in various types of cancers. The molecular mechanism underlying oncogenesis and tumor progression is still not fully elucidated, which restrict the prognostic prediction of cancer patients. Thus, it is urgent for us to identify new effective biomarkers for early diagnosis, prognosis prediction and ideal therapeutic target for cancer patients.
As a class of endogenous non coding RNA, long noncoding RNA (lncRNA) has a broad range of molecular and cellular functions, including chromatin modification, gene imprinting, alternative splicing, dosage compensation, nuclear-cytoplasmic trafficking, and inactivation of major tumor suppressor genes etc. [3,4,5]. Accumulating evidences of dysregulated lncRNAs in various cancers suggested that these greater than 200 nucleotides RNAs may contribute to cancer development and progression [6, 7]. Moreover, dramatic findings had suggested that lncRNAs may participate in a wide range of biological pathways which underlying oncogenesis [8]. Therefore, lncRNAs have attracted considerable attention as a mighty class of modulators and maybe serve as a potential biomarker for cancer patients [9,10,11,12].
LncRNA-SNHG1 (small nucleolar RNA host gene 1), located in 11q12.3, is expressed in broad ranges of cancer tissues [13,14,15,16]. Recently, emerging evidence from fundamental and clinical studies revealed that lncRNA-SNHG1 participates in tumorigenesis and exhibits poor prognostic value in different types of cancers. However, most studies reported the prognostic value of SNHG1 in cancer patients was limited by small sample size [15, 17]. Therefore, we conducted the present quantitative meta-analysis to investigate the prognostic value of SNHG1 in various cancers.
Methods
Literature search
Articles published in English and Chinese which related to the prognostic value of lncRNA SNHG1 and tumor progression were eligible for the current analysis. Up to March 20, 2018, a comprehensive search was conducted in several electronic databases: PubMed, Web of Science, Embase, ISI Web of Knowledge, Cochrane Library, BioMed Central, Springer, ScienceDirect, together with three Chinese databases: CNKI, Weipu and Wanfang. Following keywords for the online search in these databases were included: (“long noncoding RNA-” OR “noncoding RNA-” OR “lnc RNA-” OR “small nucleolar RNA host gene 1” OR “SNHG1”) AND (“carcinoma” OR “cancer” OR “tumor” OR “neoplasm”) AND (“prognosis” OR “prognostic”). The reference lists of primary publications were also manually searched to achieve potential eligible studies.
Inclusion and exclusion criteria
The following selection criteria for the eligible studies were used: 1) Definite diagnosis or histopathology confirmed for cancer patients; 2) Studies investigating the prognostic features of lncRNA SNHG1 in any malignant patients; 3) Enough information for the computation of pooled hazard ratios (HR) and 95% confidence intervals (CI). Exclusion criteria for the articles included: Studies absence of prognostic outcomes; 2) Duplicated publications; 3) Non-human research, correspondences, case reports, letters, review articles and other studies without original data.
Data extraction and quality assessment
Two authors (BZD and CZZ) carefully reviewed the information such as titles, abstracts, full texts and reference lists of each eligible article independently. The enrolled literatures were then qualified by MOOSE and PRISMA checklists (Additional file 1: Table S1 and Additional file 2: Table S2) [18]. In case that the eligible literatures only provide the data as Kaplan–Meier survival curves, the Enguage Digitizer (Version 4.1) software was used to extract the survival information from the graphical plots as the previously described method [19,20,21]. Extracted items were discussed and any contradiction was arbitrated by a third investigator (YYZ) to reach a consensus. Furthermore, the necessary elements from the enrolled articles were extracted: first author’s name; publication year; cancer resources; tumor type and stage; total cases; follow-up period; lncRNA SNHG1 detection method; cut-off values; HRs and corresponding 95% CIs.
Statistical analysis
The present meta analysis was performed with Stata SE 12.0 (Stata Corporation) and RevMan 5.3 software. The main statistical index, HRs and 95% CIs, was calculated for the aggregation of patient survival and tumor progression results. The heterogeneity between studies was determined by I2 statistics. The fixed effect model was conducted in the studies with no obvious heterogeneity (I2 < 50%) [21,22,23]. Potential publication bias was evaluated by performing Begg’s bias test and funnel plot. P value less than 0.05 was considered as statistically significant.
Results
Eligible studies
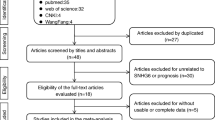
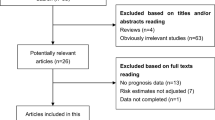
After preliminary online search, 363 literatures in total were originally retrieved from electronic databases. After duplicates removed, 350 potential articles were then subjected to abstract screened. These 278 researches which related to the lncRNA SNHG1 expression and cancer prognosis were then excluded because they do not match the inclusion criteria. Through carefully full texts assessed the remaining 72 articles, another 58 literatures were then removed according to the exclusion criteria. Ultimately, fourteen articles were enrolled in this present study. The literature screening processes were presented as a flow diagram [14,15,16,17, 24,25,26,27,28,29,30,31,32,33](Fig. 1).
Study characteristics
The main features of the enrolled 14 eligible studies included a total of 1397 participants were summarized in Table 1. Briefly, qRT-PCR was used to detect lncRNA SNHG1 expression level in all of the research studies. The detected cancer tissue samples came from neuroblastoma, esophageal squamous cell cancer, hepatocellular carcinoma, gastric cancer, epithelial ovarian cancer, osteosarcoma, lung squamous cell carcinoma, colorectal cancer, non-small cell lung cancer and lung cancer. Notably, median was selected as cut-off value in different studies. Eight of the fourteen articles focused on the association of SNHG1 with OS, PFS, EFS or RFS, and two articles investigated both OS and PFS.
Meta-analysis
Figure 2 presented the forest plot result about lncRNA SNHG1 and patient outcomes. A fix-effect model was utilized to calculate the pooled effect size because no significant heterogeneity was observed among these enrolled 10 studies (I2 = 0%). The combined results indicated that the elevated SNHG1 expression level was significantly predicted poor OS (HR = 2.06, 95% CI: 1.69–2.52, P < 0.01) and PFS (HR = 2.78, 95% CI: 1.69–4.55, P < 0.01) in various cancers. Moreover, the promoted SNHG1 level was also associated with tumor progression ((III/IV vs. I/II: HR = 1.89, 95% CI: 1.53–2.34, P < 0.01) and (III vs. I/II: HR = 1.88, 95% CI: 1.33–2.66, P < 0.01)) (Fig. 3).
a Forest plot for the association between SNHG1 expression levels with overall survival (OS). b Forest plot for the association between SNHG1 expression levels with progress free survival (PFS).c Stratified analyses for the association between SNHG1 expression with overall survival (OS). Subgroup analysis of HRs of OS by factor of cancer resources
Stratified analysis
Afterwards we set out to throw light upon the prognostic effect of SNHG1 on different cancer resources. The results from stratified analysis turned out that enforced SNHG1 expression was predictive of worse outcome in digestive system (HR = 2.04, 95% CI: 1.56–2.68, P < 0.01) and non-digestive system (HR = 2.09, 95% CI: 1.55–2.83, P < 0.01) cancer patients (Fig. 2c). No significant heterogeneity was found in the subgroup analysis.
Publication bias
To evaluate publication bias in the current meta-analysis, the indicated studies were conducted with Begg’s bias test and funnel plot analysis. The result of Begg’s test revealed the absence of significant publication bias (P = 0.474). The shape of the funnel plot was also symmetrically inverted funnels (Fig. 4).
Sensitivity analysis
Through sensitivity analysis, it was uncovered that the pooled SNHG1 HR was not significantly affected by the exclusion of any single study (Fig. 5).
Discussion
Along with the rapid expanding of high throughput genome sequencing technologies, lncRNAs were demonstrated as novel biomarkers to more precisely evaluate the prognosis of various tumors. Recently, mounting evidence suggested that over-expression lncRNA SNHG1 correlated with poor prognosis and progression of cancer patients. However, most studies reported the prognostic value of SNHG1 expression level was limited by small sample size. To the best of our knowledge, there is no systematic meta analysis concerning about lncRNA SNHG1 expression level and cancer patient outcomes.
LncRNA SNHG1, a novel lncRNA located in 11q12.3 region and containing 11 exons, which had been found significantly up-regulated in several types of cancers. The molecular mechanisms prone to be participated in the oncogenesis and progression had gradually been unveiled. For example, the dysregulation of LncRNA SNHG1 has been demonstrated to participate in Notch and Wnt/β-catenin signaling pathways in osteosarcoma and colorectal cancer [25, 34]. LncRNA SNHG1 was also suggested to interact with DNMT1, P53 and TAP63 in GC, HC and LSCC respectively [14, 15, 28]. Moreover, lncRNA SNHG1 may conduct as a competing endogenous RNA (ceRNA) that exacerbated cancer development. Up-regulated lncRNA SNHG1 reduces the miRNAs expression level, such as miR-101-3p, miR-145, miR-195, miR-326 and miR-577 in nucleus pulposus cell proliferation, osteosarcoma, hepatocellular carcinoma, non-small cell lung cancer and nasopharyngeal carcinoma respectively [17, 26, 30, 35, 36]. These encouraged evidences urged us investigating the relationship between lncRNA SNHG1 and cancer prognosis, and our analysis firstly demonstrated that high expression level of lncRNA SNHG1 was an unfavorable predictor for the clinical outcomes of various cancer patients.
Fourteen online searched studies including 1397 patients in total were pooled in this analysis, which was considered as powerful enough to consolidate our results. Several kinds of tumors, such as neuroblastoma, esophageal squamous cell cancer, hepatocellular carcinoma, gastric cancer, epithelial ovarian cancer, osteosarcoma, lung squamous cell carcinoma, colorectal cancer, non-small cell lung cancer and lung cancer, were implemented in our study. The analysis showed a pooled HR was 2.06 (95% CI: 1.69–2.52, P < 0.01), 2.78 (95% CI: 1.69–4.55, P < 0.01) and 1.89 (95% CI: 1.53–2.34, P < 0.01) for OS, PFS and tumor progression respectively. We also demonstrated that enforced SNHG1 expression was a predictor of worse outcome in digestive system (HR = 2.04, 95% CI: 1.56–2.68, P < 0.01) and non-digestive system (HR = 2.09, 95% CI: 1.55–2.83, P < 0.01) cancer patients.
Nevertheless, limitations should be refined when interpreted lncRNA SNHG1 expression level for cancer outcomes. To start with, although no publication bias was detected by statistical methods, potential bias might exist. Articles with ideal results might be published easily, which might lead to the lack of statistical power. Furthermore, the ethnicity of the cancer patients was Asian and our results may best elucidate the correlation of lncRNA SNHG1 with Asian patients.
In summary, despite some limitations mentioned above, our meta-analysis indicated that the elevated lncRNA SNHG1 level is significantly associated with cancer patients’outcome. To strengthen our results, well-designed clinical studies and multi-ethnics clinical researches should be carried out before lncRNA SNHG1 could be applied as a prognostic marker in the routine clinical guidance of cancer patients.
Conclusions
In conclusion, the present results suggest that promoted lncRNA SNHG1 expression levels are associated with OS and lncRNA SNHG1 may be used as a prognostic marker for cancer patients.
Availability of data and materials
All data analyzed during this study are included in this published article.
Abbreviations
- 95% CI:
-
95% confidence interval
- ceRNA:
-
Competing endogenous RNA
- EFS:
-
Event free survival
- HR:
-
Hazard ratio
- LncRNA:
-
Long noncoding RNA
- OS:
-
Overall survival
- PFS:
-
Progress free survival
- RFS:
-
Relapse free survival
- SNHG1 :
-
Small nucleolar RNA host gene 1
References
Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132(5):1133–45.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–38.
Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–42.
Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403.
Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–8.
Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38.
Wang J, Xu AM, Zhang JY, He XM, Pan YS, Cheng G, Qin C, Hua LX, Wang ZJ. Prognostic significance of long non-coding RNA MALAT-1 in various human carcinomas: a meta-analysis. Genet Mol Res. 2016;15(1):gmr.15017433.
Liu J, Lin J, Li Y, Zhang Y, Chen X. Prognostic role of lncRNA TUG1 for cancer outcome: evidence from 840 cancer patients. Oncotarget. 2017;8(30):50051–60.
Jing X, Lin J, Wang H, Tian L, Tian R, Zhang Y, Chen X, Zhang J. Prognostic value of lncRNA SOX2OT for Chinese cancer patients: a meta-analysis. PLoS One. 2017;12(5):e0176889.
Chen X, Lun L, Hou H, Tian R, Zhang H, Zhang Y. The value of lncRNA HULC as a prognostic factor for survival of Cancer outcome: a Meta-analysis. Cell Physiol Biochem. 2017;41(4):1424–34.
Zhang Y, Lun L, Li H, Wang Q, Lin J, Tian R, Pan H, Zhang H, Chen X. The value of lncRNA NEAT1 as a prognostic factor for survival of Cancer outcome: a Meta-analysis. Sci Rep. 2017;7(1):13080.
Chen Y, Lian YJ, Ma YQ, Wu CJ, Zheng YK, Xie NC. LncRNA SNHG1 promotes alpha-synuclein aggregation and toxicity by targeting miR-15b-5p to activate SIAH1 in human neuroblastoma SH-SY5Y cells. Neurotoxicology. 2018;68:212-21.
Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F, Li D, Yang T. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–9.
Hu Y, Ma Z, He Y, Liu W, Su Y, Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491(4):926–31.
Wang SJJ, Wang ZY, Xie YY, Wu XH. Long non-coding RNA SNHG1 is an unfavorable prognostic factor and promotes cell proliferation and migration by Wnt/β-catenin pathway in epithelial ovarian cancer. Int J Clin Exp Pathol. 2017;10(9):9284–92.
Jiang Z, Jiang C, Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/WNT/beta-catenin pathway. Biochem Biophys Res Commun. 2018;495(1):238–45.
Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis of observational studies in epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–12.
Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–34.
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Bowden J, Tierney JF, Copas AJ, Burdett S. Quantifying, displaying and accounting for heterogeneity in the meta-analysis of RCTs using standard and generalised Q statistics. BMC Med Res Methodol. 2011;11:41.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Sahu D, Hsu CL, Lin CC, Yang TW, Hsu WM, Ho SY, Juan HF, Huang HC. Co-expression analysis identifies long noncoding RNA SNHG1 as a novel predictor for event-free survival in neuroblastoma. Oncotarget. 2016;7(36):58022–37.
Zhang Y, Jin X, Wang Z, Zhang X, Liu S, Liu G. Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating notch signaling pathway in esophageal squamous cell cancer. Cancer Biomark. 2017;21(1):89–96.
Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z, Zhang F. Expression of long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Med Sci Monit. 2016;22:4820–9.
Wang J, Cao L, Wu J, Wang Q. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int J Oncol. 2018;52(1):77–88.
Zhang HY, Yang W, Zheng FS, Wang YB, Lu JB. Long non-coding RNA SNHG1 regulates zinc finger E-box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in lung squamous cell carcinoma. Biomed Pharmacother. 2017;90:650–8.
Tian T, Qiu R, Qiu X. SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget. 2018;9(2):2128–39.
Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8(11):17785–94.
Zhu Y, Li B, Liu Z, Jiang L, Wang G, Lv M, Li D. Up-regulation of lncRNA SNHG1 indicates poor prognosis and promotes cell proliferation and metastasis of colorectal cancer by activation of the Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8(67):111715–27.
Wang Q, Li Q, Zhou P, Deng D, Xue L, Shao N, Peng Y, Zhi F. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed Pharmacother. 2017;91:906–11.
Tang YMQH, Huang Y, Pan HL, Song HS, Wang X, Wu HZ, Feng ZF. LncRNA SNHG1 promotes lung Cancer cell proliferation. Chin J Biochem Mol Biol. 2017;33(12):1274–9.
Qi H, Wang J, Wang F, Ma H. Long non-coding RNA SNHG1 promotes cell proliferation and tumorigenesis in colorectal cancer via Wnt/beta-catenin signaling. Pharmazie. 2017;72(7):395–401.
Wang L, Ma X, Yan L, Wang T, Wen J, Mi G. LncRNA SNHG1 negatively regulates miR-145a-5p to enhance NUAK1 expression and promote cancer cell metastasis and invasion in nasopharyngeal carcinoma. J Cell Physiol. 2017;234:3119.
Tan H, Zhao L, Song R, Liu Y, Wang L. The long noncoding RNA SNHG1 promotes nucleus pulposus cell proliferation through regulating miR-326/ CCND1. Am J Physiol Cell Physiol. 2018;315(1):C21–7.
Acknowledgements
We are grateful to all researchers of enrolled studies.
Funding
The study was supported by Distinguished Middle-Aged and Young Scientist Encourage and Reward Foundation of Shandong Province (No. ZR2016HB08 to BZD), the National Natural Science Foundation of China (No. 81600691 to BZD), the Research and Development Project of Shandong Province (No. 2016GGB14019 to CZZ), people’s Livelihood Science and technology program of Qingdao (No.17–3–3-8-nsh to QD). The funding body was not involved in the design of the study, collection, analysis, and interpretation of data, nor the writing the manuscript. The content is solely the responsibility of the authors.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: QD and CZZ. Performed the experiments: BZD, YYZ, XC, CZZ and QD. Analyzed the data: BZD and CZZ. Contributed analysis tools/materials: BZD, YYZ, XC, CZZ and QD. Wrote the paper: BZD, QD and CZZ. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
MOOSE checklist. (DOCX 15 kb)
Additional file 2:
PRISMA checklist. (DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Dong, B., Chen, X., Zhang, Y. et al. The prognostic value of lncRNA SNHG1 in cancer patients: a meta-analysis. BMC Cancer 19, 780 (2019). https://doi.org/10.1186/s12885-019-5987-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-019-5987-4