Abstract
Background
New evidence from clinical and fundamental researches suggests that SNHG7 is involved in the occurrence and development of carcinomas. And the increased levels of SNHG7 are associated with poor prognosis in various kinds of tumors. However, the small sample size was the limitation for the prognostic value of SNHG7 in clinical application. The aim of the present meta-analysis was to conduct a qualitative analysis to explore the prognostic value of SNHG7 in various cancers.
Methods
Articles related to the SNHG7 as a prognostic biomarker for cancer patients, were comprehensive searched in several electronic databases. The enrolled articles were qualified via the preferred reporting items for systematic reviews and meta-analysis of observational studies in epidemiology checklists. Additionally, an online database based on The Cancer Genome Atlas (TCGA) was further used to validate our results.
Results
We analyzed 2418 cancer patients that met the specified criteria. The present research indicated that an elevated SNHG7 expression level was significantly associated with unfavorable overall survival (OS) (HR = 2.45, 95% CI: 2.12–2.85, p <0.001). Subgroup analysis showed that high expression levels of SNHG7 were also significantly associated with unfavorable OS in digestive system cancer (HR = 2.31, 95% CI: 1.90–2.80, p <0.001) and non-digestive system cancer (HR = 2.67, 95% CI: 2.12–3.37, p <0.001). Additionally, increased SNHG7 expression was found to be associated with tumor stage and progression (III/IV vs. I/II: HR = 1.76, 95% CI: 1.57–1.98, p <0.001). Furthermore, elevated SNHG7 expression significantly predicted lymph node metastasis (LNM) (HR = 1.98, 95% CI: 1.74–2.26, p <0.001) and distant metastasis (DM) (HR = 2.49, 95% CI: 1.88–3.30, p <0.001) respectively. No significant heterogeneity was observed among these studies. SNHG7 was significantly upregulated in four cancers and the elevated expression of SNHG7 predicted shorter OS in four cancers, worse DFS in five malignancies and worse PFI in five carcinomas based on the validation using the GEPIA on-line analysis tool.
Conclusions
The present analysis suggests that elevated SNHG7 is significantly associated with unfavorable OS, tumor progression, LNM and DM in various carcinomas, and may be served as a promising biomarker to guide therapy for cancer patients.
Similar content being viewed by others
Background
With the increasing prevalence of cancer, carcinomas had gradually been recognized as a major threaten to human health the world over [1, 2]. Although great progresses continued to be made in cancer treatment, it is not satisfactory because the long-term survival rate of many cancers is still remaining very low. With the rapid developments of science and technology, researchers gradually realize that the molecular mechanisms of tumorigenesis and development are still need further elucidated. Therefore, there is an urgent need to find new and effective clinical biomarkers for early diagnosis, prognosis and ideal therapeutic targets for cancer patients. Long noncoding RNAs (lncRNAs) have a wide range of biological functions, such as alternative splicing, chromatin modification, dosage compensation, inactivation of major tumor suppressor genes and gene imprinting etc [3,4,5].
As a modulator of biological processes, small nucleolus RNA host gene 7 (SNHG7) that located on chromosome 9q34.3, has been firstly discovered in lymphoblastoid cell lines. New evidence from clinical and fundamental researches suggests that lncRNA SNHG7 is expressed in many tissues and involved in the occurrence and development of various carcinomas. Researches have suggested that SNHG7 may associated with methylation. For example, Wu et al. had discovered that upregulated DNMT1 could induce hypermethylation of the SNHG7 promotor in hypopharyngeal cancer cells [6]. It was also reported that SNHG7 could directly bind DNMT1, which in turn binds the p53 promoter region, thus inhibiting its expression at the epigenetic level [7]. Recently, SNHG7 was discovered as differentially m6A-methylated and expressed lncRNAs in gastric cancer [8]. In cell nucleus, several proteins have been found to be regulated by SNHG7, including Bax and p21, p15 and p16, and β-catenin pathway members [9,10,11,12]. Due to the different nature of various cancer types, several signaling molecules associated with SNHG7 has gradually been unveiled, such as AKT2, BCL-2, BCRP, BDNF, CDK6, CTNNB1, Cyclin D1, DNMT1, E-cadherin, ELAVL1, ELK1, EMT, FAIM2, GALNT1, ID4, MDR1, Notch1, p15, p16, p21, PI3K/AKT/mTOR, PVT1, ROCK1, SMAD4, SOX4, SYVN1, TGF-β, WNT2B, Wnt/β-catenin etc [9, 10, 12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29].
Mounting evidences revealed that lncRNAs are deregulated in a variety of carcinomas. Therefore, lncRNAs have attracted extensive attention and may be served as potential biomarkers for carcinomas. Different studies have explored that the increase levels of lncRNA SNHG7 are associated with poor prognosis in various kinds of tumors. However, the small sample size was the limitation for the prognostic value of SNHG7 in clinical application. In the present study, a qualitative meta-analysis was conducted to explore the prognostic effect of SNHG7 on cancer patients.
Methods
Literature search and selection
Articles published in English up to Dec 30th, 2020, which related to the lncRNA SNHG7 as a prognostic biomarker for cancer patients, were comprehensive searched in several electronic databases. These databases include: Springer, Cochrane Library, Embase, BioMed Central, PubMed, Science Direct and ISI Web of Knowledge. Articles with the following keywords for the online search were included: (“SNHG7” OR “small nuclear RNA host gene 7” OR “lnc RNA-” OR “long noncoding RNA-” OR “noncoding RNA-”) AND (“neoplasm” OR “tumor” OR “cancer” OR “carcinoma”) AND (“metastasis” OR “prognosis” OR “metastatic” OR “prognostic”). Manually searched of the reference lists were also conducted to obtain potential eligible studies.
Inclusion and exclusion criteria
Inclusion criteria: 1) definite diagnosis or histopathology confirmed for carcinomas;2) studies investigating the prognostic values of lncRNA SNHG7 in various cancers; 3) sufficient information for computing pooled hazard ratios (HR) and 95% confidence intervals (CI).
Exclusion criteria: 1) duplicated articles; 2) studies absent of prognostic outcomes; 3) case reports, correspondences, letters, non-human research, review articles and other studies without original data.
Data extraction and quality assessment
After reviewed the eligible articles, two authors (YYZ and XC) extracted the necessary data independently. The necessary information from each publication was extracted: (1) last name of first author, publication year, country, cancer type, study design, stage, follow-up time and total cases; (2) SNHG7 assessment method and specimen resources; (3) hazard ratio (HR) with 95% confidence interval (CI) of SNHG7 for overall survival; (4) patient number for TNM stage and progression, lymph node metastasis and distance metastasis. Preferred reporting items for systematic reviews and meta-analyses (PRISMA) was served to qualified all of the enrolled articles (Supplementary Table S1). Enguage Digitizer (Version 4.1) software was performed to extract HRs with 95% CIs from the graphical plots if the eligible literature only provided Kaplan–Meier survival curves as the OS data [30, 31].
Validation of bioinformatics database
Gene Expression Profiling Interactive Analysis (GEPIA), a web-based tool to deliver fast and customizable functionalities based on TCGA and GTEx data (http://gepia.cancer-pku.cn/). Survival plots of the correlation between SNHG7 expression and OS or DFS were retrieved as K–M curves based on different cancer datasets from GEPIA online database. Progression free interval (PFI) analysis of various carcinomas was based on the transcriptome sequencing data from TCGA. Median was set for cutoff value. Differential expression analysis between cancer and normal tissues was conducted based on GEPIA on-line analysis. All p-value < 0.05 was regarded as significantly statistical.
Statistical analysis
The effect of SNHG7 levels on the aggregated overall survival, tumor progression, lymph node metastasis and distance metastasis were evaluated by HRs and 95% CIs. I2 statistics was used to calculated heterogeneity among the enrolled studies. The fix-effects model was performed to reveal the relationship between SNHG7 expression levels and clinical outcomes (I2< 50%) [32, 33]. Probable publication bias was evaluated by a funnel plot and Begg’s bias test [34]. A p-value < 0.05 was regarded as significantly statistical. All analyses were conducted with RevMan 5.3 software and Stata SE 12.0 (Stata Corporation).
Results
Included articles
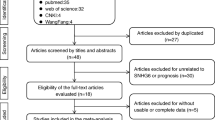
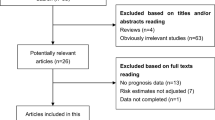
Literature screening and study selection processes were presented as Fig. S1. The preliminary online search retrieved 548 publications concerning the prognosis or metastasis of SNHG7 and cancer patients. After carefully removing the duplicates, 28 articles were excluded and 408 publications proceed to abstract screening. We then carefully remove another 385 studies according to the inclusion and exclusion criteria. Finally, 23 articles were enrolled for the meta-analysis in this study.
Characteristics of the enrolled studies
Table 1 summarized the main characteristics of the enrolled twenty-three eligible articles [6, 10, 18, 20, 21, 24, 25, 35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50]. All of the 2418 participants were from China and divided into high or low group according to the qRT-PCR or microarray results. The cut-off values were different, with median was applied in most articles. Nineteen of the enrolled studies investigated the expression level of SNHG7 and overall survival. Twenty-two articles were associated with the level of SNHG7 and tumor progression or metastasis.
Meta-analysis results
Finally, nineteen studies were enrolled to analyze lncRNA SNHG7 expression levels and cancer patient outcomes. A fix-effects model was conducted to calculate the pooled effect size because no significant heterogeneity was existed among the enrolled studies (I2 = 0%). Our results revealed that the increased SNHG7 was significantly related to the unfavorable OS (HR = 2.45, 95% CI: 2.12 – 2.85, p<0.001) (Fig. 1). Subgroup analysis showed that high expression levels of SNHG7 were also significantly associated with unfavorable OS in digestive system cancer patients (HR = 2.31, 95% CI: 1.90–2.80, p<0.001) and non-digestive system cancer patients (HR = 2.67, 95% CI: 2.12–3.37, p<0.001) (Fig. 2).
Additionally, increased SNHG7 expression was found to be associated with tumor stage and progression (III/IV vs. I/II: HR = 1.76, 95% CI: 1.57–1.98, p<0.001) (Fig. 3). Furthermore, elevated SNHG7 expression significantly predicted lymph node metastasis (LNM) (HR = 1.98, 95% CI: 1.74–2.26, p<0.001) and distant metastasis (DM) (HR = 2.49, 95% CI: 1.88–3.30, p<0.001) respectively (Fig. 4A and B).
Publication bias
Publication bias of the nineteen studies in this analysis was assessed by funnel plot and Begg’s bias test. The shape of the funnel plot was symmetrical and the Begg’s test revealed that no significant publication bias was existed (p> 0.05) (Fig. 5A).
Sensitivity analysis
Through sensitivity analysis of these nineteen enrolled articles, it was indicated that the pooled SNHG7 HR was not significantly affected by the exclusion of any single study (Fig. 5B).
Validation of the results in the GEPIA database
To further strengthen our conclusion, GEPIA on-line analysis tool was adopted to validate our results (http://gepia.cancer-pku.cn/). In terms of SNHG7 dysregulation, SNHG7 overexpression was identified in Cholangiocarcinoma (CHOL), Lymphoid Neoplasm Diffuse Large B-cell Lymphoma (DLBC), Pheochromocytoma and Paraganglioma (PCPG), and thymoma (THYM) (Fig. 6). Regarding the association between SNHG7 expression and prognosis, increased SNHG7 expression was correlated with worse OS in Adrenocortical carcinoma (ACC), Colon adenocarcinoma (COAD), Mesothelioma (MESO), Uterine Carcinosarcoma (UCS) and with worse DFS in ACC, Kidney renal papillary cell carcinoma (KIRP), Liver hepatocellular carcinoma (LIHC), Lung squamous cell carcinoma (LUSC), UCS. In addition, elevated SNHG7 associated with worse PFI in ACC, KIRP, LIHC, Prostate adenocarcinoma (PRAD) and UCS (p<0.05) (Figs. 7, 8 and 9). These results support our results and indicate that SNHG7 could be a novel prognostic biomarker for various cancers.
Discussion
Early scientists believed that lncRNAs are transcriptional noises due to the fact that most lncRNAs are generated by intron and intergenic regions of the genomes, and lack of protein coding capacity. In recent years, scientists have gradually discovered that lncRNAs may regulate the expression of target genes, involve in biological processes, and may be acted as oncogenes or tumor suppressors. With the rapid expansion of high throughput genomic sequencing technology, lncRNAs have been proved to be deregulated in various tumors, and even to be used as promising prognostic biomarkers in cancer patients. Many clinical and fundamental studies suggested that increasing levels of SNHG7 have intimate terms with unfavorable prognosis and progression in cancer patients. However, the small sample size was the limitation for the prognostic value of SNHG7 in clinical application. As far as we know, no systematic meta-analysis has been performed on SNHG7 expression levels and various cancer patients’ outcomes.
LncRNA SNHG7 has been proved to be significantly up-regulated in various carcinomas, such as bladder cancer, breast cancer, cervical cancer, colorectal cancer, gastric cancer, hepatocellular carcinoma, hypo pharyngeal cancer, melanoma, neuroblastoma, non-small cell lung cancer, pancreatic cancer, prostate cancer, thyroid cancer etc. These aberrant patterns of expression were associated with specific clinical features, such as overall survival time, lymph node metastasis, distant metastasis and TNM stage. SNHG7 serves as an oncogene and contributes to cell biological functions in various cancers, which including apoptosis inhibition, cell proliferation, cell cycle arrest, invasion, migration, and vasculogenic mimicry. Furthermore, SNHG7 may act as a competitive endogenous RNA (ceRNA) to aggravate the development of cancers. Elevated lncRNA SNHG7 may reduce the miRNAs expression level, such as miR-15a, miR-34a, miR-186, miR-193b, miR-216b, miR-342-3p, miRNA-381, miR-503, miR-514a-5p, miR-2682-5p and miR-5095 in multiple cancers [14, 19, 20, 24, 51,52,53,54]. Taken together, these articles demonstrated that SNHG7 plays an important role in tumor development and progression. These evidences encouraged us to investigate the correlation between SNHG7 expression levels and cancer prognosis. And our results demonstrated that elevated lncRNA SNHG7 is an unfavorable predictor for various cancer patients.
Twenty-three published studies that included 2418 patients were enrolled in this analysis. Our results revealed that the increased SNHG7 was significantly related to the unfavorable OS (HR = 2.45, 95% CI: 2.12 – 2.85, p< 0.001). Subgroup analysis showed that high expression levels of SNHG7 were also significantly associated with unfavorable OS in digestive system cancer (HR = 2.31, 95% CI: 1.90–2.80, p<0.001) and non-digestive system cancer (HR = 2.67, 95% CI: 2.12–3.37, p<0.001). Additionally, increased SNHG7 expression was found to be associated with tumor stage and progression (III/IV vs. I/II: HR = 1.76, 95% CI: 1.57–1.98, p<0.001). Furthermore, elevated SNHG7 expression significantly predicted lymph node metastasis (LNM) (HR = 1.98, 95% CI: 1.74–2.26, p <0.001) and distant metastasis (DM) (HR = 2.49, 95% CI: 1.88–3.30, p<0.001) respectively. GEPIA and TCGA databases were further used to validate our results as broadly as possible. High SNHG7 expression levels were observed in CHOL, DLBC, PCPG and THYM. What’s more, increased SNHG7 expression was correlated with worse OS in ACC, COAD, MESO, UCS and with worse DFS in ACC, KIRP, LUSC, UCS. In addition, elevated SNHG7 associated with worse PFI in ACC, KIRP, LIHC, PRAD and UCS. Taken together, these results indicate that SNHG7 could be a novel prognostic biomarker for various cancers.
The present meta-analysis has limitations that only the researches published in English were included. Next, this study was constrained to studies published in China, so our results may best illustrate the association between SNHG7 and Asian patients. Well-designed studies and multi-ethnic clinical researches with larger sample size should be carried out in the future. Third, some HRs are extracted by reconstructing the K-M curve, rather than directly from the original research, which would inevitably lead to possible deviations. Despite the inherent deficiencies, our study provides strong evidence that elevated lncRNA SNHG7 expression levels are prognostic for reduced OS, tumor progression, LYM and DM in cancer patients.
Conclusion
In conclusion, the present analysis implicated that elevated SNHG7 is strongly associated with OS, tumor progression, LNM and DM in carcinomas, and may be served as a promising biomarker to guide therapy for various cancer patients.
Availability of data and materials
All data analyzed during this study are included in this published article. GEPIA database is publicly available at http://gepia.cancer-pku.cn/index.html.
Abbreviations
- 95% CI:
-
95% confidence interval
- ceRNA:
-
Competing endogenous RNA
- PFI:
-
Progression free interval
- EFS:
-
Event free survival
- HR:
-
Hazard ratio
- LncRNA:
-
Long noncoding RNA
- OS:
-
Overall survival
- PFS:
-
Progress free survival
- RFS:
-
Relapse free survival
- SNHG7:
-
Small nucleolar RNA host gene 7
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30.
Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Pineros M, Znaor A, et al. Cancer statistics for the year 2020: An overview. Int J Cancer. 2021;149:778–89.
Smolarz B, Zadrozna-Nowak A, Romanowicz H. The role of lncRNA in the development of tumors, including breast cancer. Int J Mol Sci. 2021;22(16):8427–40.
Zhang D, Wu M, Xiong M, Xu C, Xiang P, Zhong X. Long noncoding RNAs: an overview. Methods Mol Biol. 2021;2372:297–305.
Camacho CV, Choudhari R, Gadad SS. Long noncoding RNAs and cancer, an overview. Steroids. 2018;133:93–5.
Wu P, Tang Y, Fang X, Xie C, Zeng J, Wang W, et al. Metformin suppresses hypopharyngeal cancer growth by epigenetically silencing long non-coding RNA SNHG7 in FaDu cells. Front Pharmacol. 2019;10:143.
Zhang GD, Gai PZ, Liao GY, Li Y. LncRNA SNHG7 participates in osteosarcoma progression by down-regulating p53 via binding to DNMT1. Eur Rev Med Pharmacol Sci. 2019;23(9):3602–10.
Lv Z, Sun L, Xu Q, Xing C, Yuan Y. Joint analysis of lncRNA m(6)A methylome and lncRNA/mRNA expression profiles in gastric cancer. Cancer Cell Int. 2020;20:464.
Xu C, Zhou J, Wang Y, Wang A, Su L, Liu S, et al. Inhibition of malignant human bladder cancer phenotypes through the down-regulation of the long non-coding RNA SNHG7. J Cancer. 2019;10(2):539–46.
Chen Y, Peng Y, Xu Z, Ge B, Xiang X, Zhang T, et al. Knockdown of lncRNA SNHG7 inhibited cell proliferation and migration in bladder cancer through activating Wnt/beta-catenin pathway. Pathol Res Pract. 2019;215(2):302–7.
Xu LJ, Yu XJ, Wei B, Hui HX, Sun Y, Dai J, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur Rev Med Pharmacol Sci. 2018;22(9):2653–61.
Wang MW, Liu J, Liu Q, Xu QH, Li TF, Jin S, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21(20):4613–22.
Cheng G, Zheng J, Wang L. LncRNA SNHG7 promotes glioma cells viability, migration and invasion by regulating miR-342-3p/AKT2 axis. Int J Neurosci. 2021;131(12):1190–202.
Deng Y, Zhao F, Zhang Z, Sun F, Wang M. Long noncoding RNA SNHG7 promotes the tumor growth and epithelial-to-mesenchymal transition via regulation of miR-34a signals in osteosarcoma. Cancer Biother Radiopharm. 2018;33(9):365–72.
She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36(5):2673–80.
She K, Yan H, Huang J, Zhou H, He J. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell Prolif. 2018;51(1):e12406.
Chen K, Abuduwufuer A, Zhang H, Luo L, Suotesiyali M, Zou Y. SNHG7 mediates cisplatin-resistance in non-small cell lung cancer by activating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2019;23(16):6935–43.
Wang YH, Huo BL, Li C, Ma G, Cao W. Knockdown of long noncoding RNA SNHG7 inhibits the proliferation and promotes apoptosis of thyroid cancer cells by downregulating BDNF. Eur Rev Med Pharmacol Sci. 2019;23(11):4815–21.
Ren J, Yang Y, Xue J, Xi Z, Hu L, Pan SJ, et al. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496(2):712–8.
Qi H, Wen B, Wu Q, Cheng W, Lou J, Wei J, et al. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother. 2018;102:326–32.
Zhong X, Long Z, Wu S, Xiao M, Hu W. LncRNA-SNHG7 regulates proliferation, apoptosis and invasion of bladder cancer cells assurance guidelines. J BUON. 2018;23(3):776–81.
Hu W, Li H, Wang S. LncRNA SNHG7 promotes the proliferation of nasopharyngeal carcinoma by miR-514a-5p/ELAVL1 axis. BMC Cancer. 2020;20(1):376.
Wang W, Chen S, Song X, Gui J, Li Y, Li M. ELK1/lncRNA-SNHG7/miR-2682-5p feedback loop enhances bladder cancer cell growth. Life Sci. 2020;262:118386.
Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9(7):722.
Cheng D, Fan J, Ma Y, Zhou Y, Qin K, Shi M, et al. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019;9:28.
Cui H, Zhang Y, Zhang Q, Chen W, Zhao H, Liang J. A comprehensive genome-wide analysis of long noncoding RNA expression profile in hepatocellular carcinoma. Cancer Med. 2017;6(12):2932–41.
Wang J, Zhang S, Li X, Gong M. LncRNA SNHG7 promotes cardiac remodeling by upregulating ROCK1 via sponging miR-34-5p. Aging. 2020;12(11):10441–56.
Zhang C, Zhu B, Li XB, Cao YQ, Yang JC, Li X, et al. Long non-coding RNA SNHG7 promotes migration and invasion of melanoma via upregulating SOX4. Eur Rev Med Pharmacol Sci. 2020;24(14):7553.
Tian F, Wang J, Zhang Z, Yang J. LncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol Res. 2020;53(1):9.
Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–34.
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Bowden J, Tierney JF, Copas AJ, Burdett S. Quantifying, displaying and accounting for heterogeneity in the meta-analysis of RCTs using standard and generalised Q statistics. BMC Med Res Methodol. 2011;11:41.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Hu Y, Wang L, Li Z, Wan Z, Shao M, Wu S, et al. Potential prognostic and diagnostic values of CDC6, CDC45, ORC6 and SNHG7 in colorectal cancer. Onco Targets Ther. 2019;12:11609–21.
Jia J, Zhang D, Zhang J, Yang L, Zhao G, Yang H, et al. Long non-coding RNA SNHG7 promotes neuroblastoma progression through sponging miR-323a-5p and miR-342-5p. Biomed Pharmacother. 2020;128:110293.
Jiang D, Wang C, He J. Long non-coding RNA DGCR5 incudes tumorigenesis of triple-negative breast cancer by affecting Wnt/beta-catenin signaling pathway. J BUON. 2020;25(2):702–8.
Li Y, Zeng C, Hu J, Pan Y, Shan Y, Liu B, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol. 2018;11(1):89.
Pang L, Cheng Y, Zou S, Song J. Long noncoding RNA SNHG7 contributes to cell proliferation, migration, invasion and epithelial to mesenchymal transition in non-small cell lung cancer by regulating miR-449a/TGIF2 axis. Thorac Cancer. 2020;11(2):264–76.
Shen A, Ma J, Hu X, Cui X. High expression of lncRNA-SNHG7 is associated with poor prognosis in hepatocellular carcinoma. Oncol Lett. 2020;19(6):3959–63.
Wang W, Liu G, Liu M, Li X. Long non-coding RNA SNHG7 promotes malignant melanoma progression through negative modulation of miR-9. Histol Histopathol. 2020;35(9):973–81.
Wu F, Sui Y, Wang Y, Xu T, Fan L, Zhu H. Long noncoding RNA SNHG7, a molecular sponge for microRNA-485, promotes the aggressive behavior of cervical cancer by regulating PAK4. Onco Targets Ther. 2020;13:685–99.
Xie Y, Wang Y, Gong R, Lin J, Li X, Ma J, et al. SNHG7 facilitates hepatocellular carcinoma occurrence by sequestering miR-9-5p to upregulate CNNM1 expression. Cancer Biother Radiopharm. 2020;35(10):731–40.
Yang X, Sun L, Wang L, Yao B, Mo H, Yang W. LncRNA SNHG7 accelerates the proliferation, migration and invasion of hepatocellular carcinoma cells via regulating miR-122-5p and RPL4. Biomed Pharmacother. 2019;118:109386.
Zeng J, Ma YX, Liu ZH, Zeng YL. LncRNA SNHG7 contributes to cell proliferation, invasion and prognosis of cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23(21):9277–85.
Zhang Y, Yuan Y, Zhang Y, Cheng L, Zhou X, Chen K. SNHG7 accelerates cell migration and invasion through regulating miR-34a-Snail-EMT axis in gastric cancer. Cell Cycle. 2020;19(1):142–52.
Zhang P, Shi L, Song L, Long Y, Yuan K, Ding W, et al. LncRNA CRNDE and lncRNA SNHG7 are promising biomarkers for prognosis in synchronous colorectal liver metastasis following hepatectomy. Cancer Manag Res. 2020;12:1681–92.
Zhao D, Zhang H, Long J, Li M. LncRNA snhg7 functions as an oncogene in cervical cancer by sponging miR-485-5p to modulate JUND expression. Onco Targets Ther. 2020;13:1677–89.
Chi R, Chen X, Liu M, Zhang H, Li F, Fan X, et al. Role of SNHG7-miR-653-5p-STAT2 feedback loop in regulating neuroblastoma progression. J Cell Physiol. 2019;234(8):13403–12.
Xia Q, Li J, Yang Z, Zhang D, Tian J, Gu B. Long non-coding RNA small nucleolar RNA host gene 7 expression level in prostate cancer tissues predicts the prognosis of patients with prostate cancer. Medicine. 2020;99(7):e18993.
Han Y, Hu H, Zhou J. Knockdown of LncRNA SNHG7 inhibited epithelial-mesenchymal transition in prostate cancer though miR-324-3p/WNT2B axis in vitro. Pathol Res Pract. 2019;215(10):152537.
Gao YT, Zhou YC. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast cancer progression by sponging miRNA-381. Eur Rev Med Pharmacol Sci. 2019;23(15):6588–95.
Zhang H, Zhang XY, Kang XN, Jin LJ, Wang ZY. LncRNA-SNHG7 enhances chemotherapy resistance and cell viability of breast cancer cells by regulating miR-186. Cancer Manag Res. 2020;12:10163–72.
Li Y, Guo X, Wei Y. LncRNA SNHG7 inhibits proliferation and invasion of breast cancer cells by regulating miR-15a expression. J BUON. 2020;25(4):1792–8.
Acknowledgements
We are grateful to all researchers of enrolled studies.
Funding
This study is fully supported by the National Natural Science Foundation of China (No. 81601821 to YYZ), the Natural Science Foundation of Shandong Province (No. ZR201910230488, to XC), the Natural Science Foundation of Shandong Province (No. ZR202102180776, to YYZ), Livelihood Science and technology program of Qingdao (No.17-3-3-8-nsh to QD), Shandong Key Laboratory of Digital Medicine and Computer-Assisted Surgery Foundation (to QD).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: X.C, Y.Y.Z. and Q.D. Performed the experiments: X.C., Y.Y.Z., Q.W.T., SFH., H.M.W. and Q.D. Analyzed the data: X.C., Q.W., Q.W.T. and Y.Y.Z. Contributed reagents/materials/analysis tools: X.C., Q.D. and Y.Y.Z. Wrote the paper: X.C. and Y.Y.Z. The author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
: Figure S1. Flow diagram of the study search and selection process.
Additional file 2
: Table S1. PRISMA checklist.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, Y., Tian, Q., Huang, S. et al. Prognostic effect of lncRNA SNHG7 on cancer outcome: a meta and bioinformatic analysis. BMC Cancer 22, 10 (2022). https://doi.org/10.1186/s12885-021-09068-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-021-09068-w