Abstract
Background
Uridine disphosphate (UDP) glycosyltransferases (UGTs) act upon a huge variety of highly diverse and complex substrates, such as phytohormones and specialized metabolites, to regulate plant growth, development, disease resistance, and environmental interactions. However, a comprehensive investigation of UGT genes in tobacco has not been conducted.
Results
In this study, we carried out a genome-wide analysis of family-1 UDP glycosyltransferases in Nicotiana tabacum. We predicted 276 NtUGT genes, which were classified into 18 major phylogenetic subgroups. The NtUGT genes were invariably distributed among all the 24 chromosomes with structural diversity in exon/intron structure, conserved motifs, and cis-acting elements of promoters. Three groups of proteins which involved in flavonoid biosynthesis, plant growth and development, transportation and modification were identified that interact with NtUGT proteins using the PPI analysis. Expression analysis of NtUGT genes in cold stress, drought stress and different flower color using both online RNA-Seq data and the realtime PCR analysis, suggested the distinct role of NtUGT genes in resistance of cold, drought and in flavonoid biosynthesis. The enzymatic activities of seven NtUGT proteins that potentially involved in flavonoid glycosylation were analyzed, and found that all seven exhibited activity on myricetin; six (NtUGT108, NtUGT123, NtUGT141, NtUGT155, NtUGT179, and NtUGT195) showed activity on cyanidin; and three (NtUGT108, NtUGT195, and NtUGT217) were active on the flavonol aglycones kaempferol and quercetin, which catalyzing the substrates (myricetin, cyanidin or flavonol) to form new products. We further investigated the enzymatic products and enzymatic properties of NtUGT108, NtUGT195, and NtUGT217, suggested their diverse enzymatic activity toward flavonol, and NtUGT217 showed the highest catalyzed efficient toward quercetin. Overexpression of NtUGT217 significantly increase the content levels of the quercetin-3-O-glucoside, quercetin-3-O-rutinoside and kaempferol-3-O-rutinoside in transgenic tobacco leaves.
Conclusion
We identified 276 UGT genes in Nicotiana tabacum. Our study uncovered valuable information about the phylogenetic structure, distribution, genomic characters, expression patterns and enzymatic activity of NtUGT genes in tobacco. We further identified three NtUGT genes involved in flavonoid biosynthesis, and overexpressed NtUGT217 to validate its function in catalyze quercetin. The results provide key candidate NtUGT genes for future breeding of cold and drought resistance and for potential metabolic engineering of flavonoid compounds.
Similar content being viewed by others
Background
Glycosylation is an important modification reaction that plays crucial roles in plant growth and in responses to biotic and abiotic stresses [1]. Glycosyltransferases (GTs) facilitate glycosylation by catalyzing the transfer of sugar molecules from activated donors to specific receptors [2]. Family-1 GTs, usually referred to as uridine disphosphate (UDP) glycosyltransferases (UGTs), are the most common GTs in plants and have significant effects on plant growth and development. UGT proteins possess a highly conserved consensus sequence near the C-terminal that is 44 amino acids (aa) in length and is referred to as the plant secondary product glycosyltransferase (PSPG) box [3]. An increasing number of putative UGT-encoding genes have been identified in plants, including 107 in Arabidopsis thaliana, 130 in Prunus mume, 147 in maize, 179 in wheat, 180 in rice, 181 in grape, 182 in soybean, and 241 in apple [4-9]. Diverse multi-gene UGT family members function together to modulate complicated biochemical processes in plant cells, which in turn affects numerous biological activities and functions.
UGT proteins catalyze the conversion of sugar groups from UDP activated sugars to substrates such as hormones and specialized metabolites, and are therefore involved in biosynthesis of natural plant products such as flavonoids, terpenoids, steroids, and hormones, which regulate plant growth, development [10, 11]. However, only a few UGT proteins have been documented. For example, 26 UGT proteins from the UGT71, UGT73, UGT74, UGT85, UGT91, and UGT94 families in multiple plants have been functionally characterized as catalyzing triterpene glycosylation [12]. UGT79B1 and UGT91A1 in Arabidopsis, strawberry, peach, and kiwifruit were reported to mediate anthocyanin modification [13-15]. UGT13, UGT72AD1, UGT72AF1, UGT72AH1, UGT72V3, UGT72Z2, UGT73A20/24/25, UGT73C20, UGT73F2, UGT76F1, UGT78A14, UGT78D1/2/3, UGT80B1, UGT80A2, UGT88A1, UGT88E14/15/16/18/19, UGT92G4, UGT91Q2, and UGT716A1 have been isolated and confirmed to function in catalyzing flavonoid glycosylation [16-21]. In Arabidopsis, 118 UGT genes were shown to be differentially expressed in response to treatment with 2,4-dichlorophenoxyacetic acid and trichostatin, abscisic acid (ABA) and salicylic acid (SA), indole acetic acid (IAA), methyl jasmonate (MeJA), or zeatin [11]. UGT71B6, UGT71B7, and UGT71B8 play crucial roles in ABA homeostasis and in adaptation to various abiotic stresses [22]. UGT84B1 and UGT74D1 modulate IAA levels throughout plant development by dual IAA and oxIAA glycosylation [23]. UGT72AD1 and UGT72Z2 overexpression results in significant inhibition of soybean root growth, suggesting a role of these genes in auxin homeostasis [19].
UGTs not only function as glycosylate acceptor molecules, but also play pivotal roles in biotic and abiotic stress responses. They function in stabilizing and enhancing water solubility, inactivating or detoxifying natural products [24], promoting regulation of metabolic homeostasis and detoxifying exogenous substances [3]. TaUGT2, TaUGT3, TaUGT4, and TaUGT1287 were reported associated with scab resistance [25, 26]. Down-regulation of CsUGT91Q2 and CsUGT78A14 reduces cold-stress tolerance [27, 28]. AtUGT76b1 knockout mutants have a dwarf phenotype and a constitutive Pseudomonas infection defense response [29]. In rice, UGT90A1 and IAGT1 are significantly up-regulated in response to a combined hydroxyurea and IAA treatment, suggesting that these genes activate auxin-glucose conjugation to protect rice seedlings against hydroxyurea-induced phytotoxicity [30]. In this study, we aimed to identify putative UGT genes in tobacco and to further explore NtUGT genes that may be involved in abiotic stress resistance and flavonoid biosynthesis. We also aimed to characterize the enzymatic properties of recombinant NtUGT proteins to provide a basis for further functional analyses in the future.
Results
Identification of NtUGT genes in N. tabacum
Complete sequencing and de novo genome assembly of the common tobacco cultivar K326 has greatly facilitated the identification of tobacco gene families. Using the latest release of the tobacco genome, we used HMMER 3.0 and the HMM profile of the conserved UGT domain to search N. tabacum for putative UGT genes. After removing genes encoding redundant proteins and sequences that were too short or too divergent, 276 candidate UGT proteins were identified for further analysis (GenBank accession numbers: OP616116-OP616391). All candidates were between 98 and 1443 aa in length and contained the signature PSPG motif. All of the identified UGTs contained two major domains, a conserved C-terminal domain and a variable N-terminal domain, although the overall sequence diversity was high between genes. The NtUGT genes were named based on their chromosomal location (or scaffold location, where necessary). The predicted subcellular locations of the NtUGT genes varied widely, although most were predicted to be localized to the chloroplast (129). Seventy-five NtUGTs were predicted to be located in the cytoplasm, and 37 were predicted to be located in the nucleus (Supplementary Table S4 and Figure S1).
GO term analysis of all 276 putative NtUGT genes showed that they were associated with 15 molecular function terms and 23 biological process terms (Supplementary Figure S2). Most of the putative NtUGT genes were annotated as being involved in flavonoid biosynthesis, flavonoid metabolic processes, and various flavonoid glucuronidation processes (Supplementary Figure S2A). The putative NtUGT genes were associated with 20 major KEGG biochemical pathways related to general and specialized metabolic processes (Supplementary Figure S2B and Table S5), including starch and sucrose metabolism, sphingolipid metabolism, ether lipid metabolism, flavonoid biosynthesis, phenylpropanoid biosynthesis, carotenoid biosynthesis, steroid hormone biosynthesis, zeatin biosynthesis, ascorbate and aldarate metabolism, drug metabolism, retinol metabolism, chemical carcinogenesis, metabolism of xenobiotics by cytochrome P450, porphyrin and chlorophyll metabolism, glucosinolate biosynthesis, cyanoamino acid metabolism, pentose and glucuronate interconversions, and tryptophan metabolism.
Phylogenetic analysis of NtUGT proteins
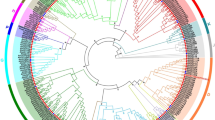
The relationships between the 276 tobacco and 114 Arabidopsis UGT proteins identified were next analyzed. Based on the protein sequences of typical Arabidopsis UGTs in each group, the phylogenetic tree constructed from these 390 proteins showed 18 major subgroups, namely 14 subgroups (A–N) that were present in all higher plants [4, 5], one O subgroup that was present in some higher plant species such as maize [32], and three out-group subgroups (OG1-OG3), which not belong to present known subgroups. (Fig. 1). Subgroup O had the most members (40 proteins), followed by subgroup E (37 proteins). Statistical analysis of the distribution of NtUGT protein functions showed that nearly all proteins with the same function were present in the same subgroup. As reported UGTs in Arabidopsis, Brassica species, maize and soybean, UGT proteins related to polyphenolic and flavonoid synthesis always enriched in subgroup A, E, F, L and M [4, 11, 13-18], proteins related to terpene metabolism were enriched in subgroups D and M [12, 28], proteins related to plant hormones were enriched in subgroups H, L, K, and O [11, 19, 22, 23], and proteins related to zeatin metabolism were almost enriched in subgroup O [5].
Phylogenetic analysis of UGT family members in Nicotiana tabacum. ClustalW and MEGA7 were used for alignment of the full-length sequences of 276 tobacco and 114 Arabidopsis UGTs and for phylogenetic tree construction. A green triangle at the end of a branch indicates an Arabidopsis gene whereas a red circle indicates a tobacco gene
Chromosomal distribution of NtUGT genes
Using the annotation information for NtUGT genes from the N. tabacum genome database, the chromosomal locations of NtUGT genes were illustrated to determine NtUGT distribution in tobacco. Due to the relatively low assembly quality of the N. tabacum genome, only 123 NtUGT genes could be mapped to chromosomes. The mapped NtUGT genes were unevenly distributed across 24 chromosomes. The number of NtUGT genes varied between 1 and 15 per chromosome, with relatively high density observed on chromosomes 17, 13, 15, and 24 (Supplementary Figure S3). All NtUGT genes located in chromosome 17 were members of subgroup E. Tandem duplication events were also analyzed to determine the importance of duplication events in shaping chromosomes and causing tandem exons in NtUGT genes. To analyze the syntenic relationships of NtUGTs, we mapped the 18 pairs of NtUGTs derived from tandem duplication to the duplicated blocks (Supplementary Figure S3). Eighteen gene pairs among 35 NtUGT genes were derived from tandem duplication. Interestingly, gene pairs derived from tandem duplication were each present in the same subgroup, and most of the gene pairs had the same function (Table 1). This suggested that tandem duplication may have played a major role in expansion of the NtUGT gene family. Ka/Ks analysis revealed that the 18 pairs of genes were under selective pressure; eight were under positive selection, whereas the other 10 pairs were under purifying selection (Table 2).
Genomic characteristics of NtUGT genes
We analyzed the exon/intron structure, conserved motifs, and cis-acting elements in NtUGT promoters to investigate the structural diversity among these genes. We found that exon number was not evenly distributed. There were 109 NtUGT genes with no introns and 161 with at least one intron. Of the 161 intron-containing NtUGT genes, 115 possessed one intron, 33 had two introns, and the remaining genes possessed between three and 16 introns (Supplementary Figure S4). NtUGT140 had the largest number of exons and the most complex gene structure, with 17 exons and 16 introns. In the phylogenetic tree, most NtUGTs with the same number of introns clustered together. Most of the intron-less NtUGTs were clustered in subgroups O (22 intron-less NtUGTs), OG3 (20), D (20), E (13), and A (11). NtUGTs with single intron were primarily clustered in subgroups L, E, O, H, A, G, OG3, and D, the latter of which contained 26 single-intron genes. NtUGT genes with two introns were mainly clustered in subgroups E (seven), OG3 (six), and D (five). In general, members of each subgroup exhibited similarity in intron/exon characteristics (Supplementary table S6).
During the long evolutionary history of plants, gene expression has been precisely regulated by transcription factors. Cis-elements in gene promoter sequences determine which transcription factors bind to a gene to regulate expression. In this study, we conducted a detailed investigation of all cis-elements present in the promoter regions (classified as 1500 bp upstream of the transcription start site) of UGT family genes. We found a total of 4472 cis-elements of 63 different types (Fig. 2A). The most abundant were MYB-binding sites (MBSs) (20%), ABREs (12%), LTRs (11%), TGACG-motifs (10%), MYCs (9%), G-boxes (5%), GT1-motifs (4%) and W boxes (4%) (Fig. 2B). MBSs were associated with drought stress, light responsiveness, and flavonoid biosynthesis regulation. ABREs were involved in responses to abiotic stress and light. G-boxes and GT1-motifs were involved in light responsiveness; LTRs were associated with low-temperature responsiveness; MYCs were involved in light responsiveness and temperature responsiveness; TGACG-motifs were involved in MeJA responsiveness; and W boxes were associated with SA responsiveness. We also identified other cis-elements that were associated with responses to gibberellin (namely the TATC-box, GARE-motif, and P-box), auxin (the TGA-element), and light (the GATA-motif, MRE, TCT-motif, ATCT-motif, chs-CMA1a, chs-CMA2a, and ACE). These results indicated that NtUGT gene expression may be influenced by a wide range of developmental processes and environmental factors.
To further analyze the distribution of major cis-elements in NtUGT genes, the type and number of cis-elements in each subgroup were counted and described in detail (Supplementary Figure S5). MBSs were the most abundant type of cis-element in most of the subgroups, except in subgroups B, C, and F; in those three subgroups, the most abundant types were ABREs, G-boxes, and MYCs, respectively. Subgroup E contained the largest number of cis-element types (50), followed by subgroups L and O (49 each).
NtUGT protein–protein interaction (PPI) networks
Mining the proteins that interact with NtUGTs can aid in understanding NtUGT functions. Using NtUGTs as bait proteins revealed a total of three groups of interacting proteins. Group I comprised 25 NtUGTs that interacted with each other (red nodes) and with other types of proteins (blue nodes) (Fig. 3). NtUGT60, NtUGT94, NtUGT62, NtUGT170, and NtUGT192 were found to interact with five flavonoid biosynthesis related structure genes, including Nitab4.5_0000027g0470 (chalcone isomerase-like, CHI), Nitab4.5_0001066g0070 (chalcone synthase, CHS), Nitab4.5_0001410g0070 (flavonoid 3'-monooxygenase), Nitab4.5_0010547g0010 (leucoanthocyanidin dioxygenase, LDOX), Nitab4.5_0000178g0360 (dihydroflavonol 4-reductase, DFR), and Nitab4.5_0005357g0020 (anthocyanidin synthase, ANS),which suggested that NtUGT60, NtUGT94, NtUGT62, NtUGT170, and NtUGT192 may be involved in flavonoid metabolism and have important roles in flavonoid glycosylation. Meanwhile, NtUGT60 and NtUGT192 were found also interact with Nitab4.5_0013249g0010 (CYP77B1), a fatty acid epoxygenase specific to flowering plant [33]. NtUGT94 was found also interact with Nitab4.5_0002221g0060 (ferulate-5-hydroxylase, F5H) and Nitab4.5_0007775g0010 (caffeic acid 3-O-methyltransferase, COMT), which was important structure genes involved in lignin biosynthesis. NtUGT60, NtUGT192 and NtUGT94 may be function in both flavonoid biosynthesis and in other metabolism. The results suggested that some NtUGTs have broad substrates and function in various biological process. In Group II, NtUGT220 was found to interact with 11 proteins, including Nitab4.5_0006530g0020 (pentatricopeptide repeat protein, PPR), Nitab4.5_0008322g0010 (RNA binding domain of NusB), Nitab4.5_0009676g0020 (homeobox transcription factor, ISS), Nitab4.5_0002331g0050 (ribonuclease III C terminal domain), Nitab4.5_0002901g0080 (transcription-repair coupling factor), Nitab4.5_0016936g0020 (RNA recognition motif) and Nitab4.5_0006338g0020 (which contains a DNA-binding motif found in homing endonucleases), which were associated with DNA replication, transcription, and translation. We therefore hypothesized that NtUGT220 may play an important role in plant growth and development. In Group III, NtUGT245 was found to interact with three proteins: Nitab4.5_0002978g0060 (potassium transporter), Nitab4.5_0007141g0030 (serine/threonine-protein kinase fray), and Nitab4.5_0007879g0060 (protein furry homolog-like). These proteins may be associated with the function of transportation and modification.
NtUGT protein–protein interaction (PPI) network. Each node in the PPI network represents all proteins generated by the associated single gene. Node size represents the degree of interaction and edge thickness represents the strength of the interaction between two proteins. Nodes representing NtUGTs are red whereas those representing non-NtUGT proteins that interact with NtUGTs are blue
Differential NtUGT gene expression under low-temperature and drought stress
Previously published tobacco abiotic stress datasets were analyzed. Using thresholds of |log2(FoldChange)|≥ 1 and p ≤ 0.05, 92 NtUGTs were classified as differentially expressed genes (DEGs) under low-temperature stress.28 NtUGT genes were down-regulated and 64 NtUGT genes were up-regulated in response to low-temperature stress. Among them, NtUGT46, NtUGT54, NtUGT56, NtUGT103, NtUGT107, NtUGT113, NtUGT117, NtUGT183, NtUGT242, NtUGT265, and NtUGT269 were significantly down-regulated; NtUGT90, NtUGT108, NtUGT124, NtUGT144, NtUGT156, NtUGT179, and NtUGT258 were significantly up-regulated; NtUGT18, NtUGT43, NtUGT123, NtUGT188, NtUGT232 and NtUGT232 were specially induced expressed under cold stress (Fig. 4A). The above results indicated that the significant different expressed NtUGT genes might play roles in regulating tobacco resistance to low temperature stress. Under drought stress, 65 NtUGTs were identified as DEGs. Interestingly, under drought stress, most NtUGTs were down-regulated, and 22 NtUGT genes were significantly down-regulated; only NtUGT208, NtUGT192, NtUGT202, NtUGT98, and NtUGT131 were up-regulated, and only NtUGT98 was significantly up-regulated (Fig. 4B). There were 38 NtUGT genes that were significantly differentially expressed in response to both cold and drought stress. Among them, NtUGT98, NtUGT205, and NtUGT208 were up-regulated under both conditions, 17 NtUGT genes were down-regulated under stress conditions compared with the control, and the remaining 18 NtUGT genes were up-regulated under cold stress while down regulated under drought stress (Fig. 4).
Significantly differentially expressed NtUGT genes in response to cold and drought stress. A NtUGT genes differentially expressed in response to cold stress. B NtUGT genes differentially expressed in response to drought stress. NtUGT genes were classified as differentially expressed using threshold values of |log2(FoldChange)|≥ 1 and p ≤ 0.05
Differential NtUGT gene expression between white and pink flowers
Using thresholds of |log2(FoldChange)|≥ 1 and p ≤ 0.05, 26 NtUGTs were identified as DEGs between flower tissues of two different colors (Fig. 5). Compared with white flowers (WF), a total of 12 NtUGT genes were significantly up-regulated in pink flowers (YCK), including NtUGT195, NtUGT123, NtUGT72, NtUGT86, NtUGT146, NtUGT149, NtUGT167, NtUGT224, NtUGT263, NtUGT266, and NtUGT2. In contrast, 14 NtUGT genes were significantly up-regulated in WF compared to YCK, including NtUGT20, NtUGT60, NtUGT94, NtUGT108, NtUGT127, NtUGT141, NtUGT155, NtUGT179, NtUGT196, NtUGT220, NtUGT232, NtUGT245, NtUGT237, and NtUGT251.
12 NtUGT genes were differentially expressed in both cold-treated plants and between differently-colored flowers. Among them, NtUGT2, NtUGT72, NtUGT123, NtUGT149, NtUGT195, NtUGT224, and NtUGT263 were up-regulated both in cold stress and in YCK, whereas NtUGT108, NtUGT179, and NtUGT232 were up-regulated both in cold stress in WF. 7 NtUGT genes were differentially expressed in both under drought stress and between differently-colored flowers. NtUGT149, NtUGT263, and NtUGT266 were down-regulated both in drought stress and in WF, whereas NtUGT108, NtUGT127, NtUGT245, and NtUGT251 were down-regulated both in drought stress in YCK. Five identical NtUGT genes (NtUGT108, NtUGT149, NtUGT245, NtUGT263, and NtUGT266) were differentially expressed in low-temperature stress, drought stress, and differently-colored flowers.
qRT-PCR validation of DEGs
qRT-PCR analysis was used to confirm the expression patterns of nine randomly-selected NtUGTs identified as DEGs based on the RNA-Seq data. The qRT-PCR results showed that NtUGT88, NtUGT108, NtUGT123, NtUGT179, NtUGT195, and NtUGT140 were significantly up-regulated under cold stress compared to control plants, which almost consist with the expression of NtUGT88, NtUGT108, NtUGT123, NtUGT179, and NtUGT195 that were detected up-regulated in RNA-Seq data. Under drought stress, NtUGT86, NtUGT227, and NtUGT140 were detected up-regulated, whereas NtUGT108 was down-regulated both in qRT-PCR analysis and in RNA-Seq analysis (Fig. 4 and Figure S6A). Meanwhile, NtUGT108, NtUGT179, and NtUGT141 were up-regulated and NtUGT86, NtUGT123, and NtUGT195 were down-regulated in WF both in qRT-PCR analysis and in RNA-Seq analysis. (Fig. 5 and Figure S6B). The expression trends of these nine genes were thus consistent with the RNA-Seq data, validating the utility of the transcriptome data.
Substrate specificity of seven recombinant NtUGT proteins
To investigate the enzymatic activity of NtUGT proteins predicted to function in flavonoid biosynthesis, seven candidate NtUGT genes with differential expression between white and pink flowers were selected for further enzymatic analysis. The seven candidate NtUGT genes were cloned and expressed in E. coli. The recombinant proteins were then evaluated in enzymatic assays using four flavonoid aglycones as sugar acceptors and UDP-glucose as the initial sugar donors. The enzymatic products of the seven NtUGTs were identified via HPLC. We found that all seven NtUGTs had activity on myricetin, and six NtUGTs (NtUGT108, NtUGT123, NtUGT141, NtUGT155, NtUGT179, and NtUGT195) showed activity on cyanidin. Three NtUGTs (NtUGT108, NtUGT195, and NtUGT217) also showed activity on flavonol aglycones, namely kaempferol and quercetin (Table 3). Based on the authentic reference standards, the enzymatic products of NtUGT108, NtUGT195, and NtUGT217 had flavonol monoglucosides at different OH groups (3, 4, and 7) (Fig. 6). These were identified as kaempferol 3-O-glucoside (K3G), quercetin 3-O-glucoside (Q3G), quercetin 4-O-glucoside (Q4G), and kaempferol 7-O-glucoside (K7G). NtUGT217 was identified as having much higher enzymatic activity on flavonols, ~ tenfold higher than NtUGT108 and NtUGT195 had. The products of quercetin catalyzed by NtUGT108 and NtUGT195 were identified as mainly Q4G with some Q3G, whereas NtUGT217 produced primarily Q3G (Fig. 6). NtUGT108, NtUGT195, and NtUGT217 primarily catalyzed formation of K3G (and small amounts of K7G) from kaempferol. These results strongly suggested that NtUGT108, NtUGT195, and NtUGT217 acted on flavonol, with strict regio-specificity at the 3-, 4-, and 7-OH positions, respectively.
Analysis of the enzymatic reaction products of three recombinant NtUGT proteins. A–D Representative HPLC chromatograms of the products formed by action of the following NtUGT proteins on quercetin: (A) control (no enzyme); (B) NtUGT108; (C) NtUGT195; and (D) NtUGT217. E–H Representative HPLC chromatograms of the products formed by action of NtUGT proteins on kaempferol: (E) control (no enzyme); (F) NtUGT108; (G) NtUGT195; and (H) NtUGT217. Q, quercetin (green arrow); Q3G, quercetin 3-O-glucoside (purple arrow); Q4G, quercetin 4-O-glucoside (red arrow); K, kaempferol (blue arrow); K3G, kaempferol 3-O-glucoside (orange arrow); Q7G, kaempferol 7-O-glucoside (black arrow)
In order to elucidate the enzymatic properties of three flavonol aglycone related recombinant NtUGT proteins (NtUGT108, NtUGT195, and NtUGT217), their enzyme kinetic parameters were further determined with UDP-glucose as substrate and compared with each other. The results showed that three NtUGTs had different Km values for quercetin 3-O-glucoside (Q3G), quercetin 4-O-glucoside (Q4G), kaempferol 3-O-glucoside (K3G) and kaempferol 7-O-glucoside (K7G) (Table 4). For quercetin as substrate, NtUGT217 got the lowest Km value (1.19 × 10–13 mM) and the high Kcat/Km value (541,044,022.1 s-1 M-1) for produce quercetin 3-O-glucoside (Q3G), indicating NtUGT217 were the most production efficient quercetin 3-O-glucoside than other than NtUGT108 and NtUGT195. While NtUGT108 showed higher activity toward quercetin 4-O-glucoside (Q4G) than Q3G, NtUGT195 showed the low activity toward Q4G and Q3G. For Kaempferol, three NtUGT proteins showed higher activity toward kaempferol 3-O-glucoside (K3G) than kaempferol 7-O-glucoside (K7G).
NtUGT217 overexpression in N. tabacum
To evaluate the functions of NtUGT genes in flavonol biosynthesis in vivo, we overexpressed NtUGT217 in tobacco via Agrobacterium-mediated transformation. Three transgenic lines with high transcript levels (U217OE-3, U217OE-9, and U217OE-15) were detected by qRT-PCR (Fig. 7B) and analyzed in detail. The flavonols in tobacco leaves were quercetin-3-O-rutinoside and kaempferol-3-O-rutinoside. We detected increased levels of both of those two compounds and quercetin-3-O-glucoside in all three transgenic NtUGT217-overexpression tobacco lines (Fig. 7C). Subcellular localization analysis showed that NtUGT217 was localized to both the nucleus and cytoplasm (Fig. 7A). This was consistent with the localization of nearly all structural proteins involved in flavonoid biosynthesis, such as CHS, CHI, and FLS. Cellular co-localization of those proteins provided potential possibility of protein–protein interactions and functioning in flavonoid biosynthesis. Above all, the results suggested that NtUGT217 may have important roles in flavonol glycosylation.
Flavonol glucoside contents in transgenic tobacco overexpressing NtUGT217. A Subcellular localization analysis of NtUGT217 gene. Left, confocal micrograph showing green fluorescent protein (GFP) fluorescence; middle, the corresponding differential interference contrast (bright field) image; right, merged fluorescent and bright field image. Scale bars = 20 μm. B Expression levels of NtUGT217 in transgenic tobacco lines overexpressing NtUGT217. C Levels of quercetin-3-O-glucoside, quercetin-3-O-rutinoside and kaempferol-3-O-rutinoside were dramatically increased in NtUGT217-overexpression lines
Discussion
Multi-gene UGT families have been identified in several plant species. However, to our knowledge, no further information has been reported about the UGT gene family in N. tabacum. In this study, we sought to identify and determine putative functions of all UGT genes in tobacco. In total, we identified 276 putative NtUGT genes (Fig. 1), which encoded proteins of a large range of amino acids lengths. The intron numbers of the 276 NtUGT family members varied from 0 to 16, and the gene structure was shown to be complex. Of the 276 putative NtUGT genes, 38% lacked introns, which was lower than the rate of 58%, 55%, and 60% of genes lacking introns in Arabidopsis, flax, and maize, respectively [31, 32]. Phylogenetic analysis revealed that the 276 NtUGTs were distributed among 18 subgroups, namely 14 conserved subgroups (subgroups A–N), which were present in all species [5]; one newly discovered subgroup (subgroup O), which was found in some higher plant species such as apple, peach, poplar, and maize [33]; and three out-group subgroups (OG1-OG3), which were not subject to known subgroups. Subgroup O contained the most NtUGT genes (40), accounting for ~ 14% of all NtUGT genes identified in tobacco. Expansion of this subgroup in tobacco indicated that they have performed vital functions associated with tobacco evolution, and their absence in Arabidopsis indicates they were lost during the evolution of Arabidopsis or that the family expanded after separation from the last common ancestor of tobacco and Arabidopsis. The new subgroup OG3 contained the third highest number of NtUGT genes (33), indicating their indispensable functions specific to speciation (i.e., formation, adaption, and development) of N. tabacum. Expansions of subgroups A, C, D, E, G, I, J, K, L, M, and N in tobacco compared to Arabidopsis indicated that multiple functions were associated with these subgroups of UGTs, and they had broad substrate specificity. Subgroups B and F were not found to be expanded, suggesting that they had conserved substrate specificity (Fig. 1). Surprisingly, subgroup H showed a decreased number of NtUGTs in tobacco compared to Arabidopsis, with a shrinking of the gene family, which indicated that these NtUGTs were partially lost during evolution, which may have limited their functions in tobacco (Fig. 1). Formation, expansion, conservation, and reduction of NtUGT genes in each phylogenetic subgroup reflect the evolutionary challenges that plants must overcome to survive.
In multiple plant species, UGTs have been reported to respond to various abiotic stress conditions, including low temperature, drought, and high salt [11, 34]. In this study, gene expression was analyzed to better understand the roles of tobacco NtUGT genes in flavonoid biosynthesis and in drought and cold stress resistance. The promoter sequences of NtUGTs were shown to contain many cis-elements related to responses to abiotic stressors (such as light, temperature, and drought) and to hormones and flavonoid biosynthesis, suggesting that NtUGTs may be involved in responses to a range of stressors. An increasing number of UGT genes have been shown to function in cold resistance in multiple plants, including AtUGT79B2 and AtUGT79B3 [35], TaUGT91Q2 [28], EjUGT92 and EjUGT88 [36], and MdUGT83L3 [37]. Under cold stress, we identified 26 significantly up-regulated NtUGT genes and eight that were significantly down-regulated. Cold stress response elements such as ABRE, DRF, W-box, ARE, G-box were present in the promoter fragments of those significant different expressed NtUGT genes under cold stress, which suggested their potential roles in cold resistance.
UGT genes have also been shown to function in drought resistance, include OsUGT85E1 [38], AhUGT71K1 and AhUGT73B4 [39], UGT003 and UGT024 in alfalfa (Medicago sativa) [40], AtUGT74E2 [41], AtUGT71C5 [42], AtUGT76E11 [43], and ZmUGT2 [44]. Interestingly, of the 30 NtUGT genes significantly differentially expressed under drought stress, the majority (25) were significantly down-regulated; and five NtUGT genes were significantly up-regulated (Fig. 4B). The expression patterns of NtUGTs were different compared to MsUGTs, most of which were significantly up-regulated under drought stress [40]. RNA interference of UGT75C1 in Solanum lycopersicum reportedly enhances the ability of transgenic tomatoes to resist drought stress [45]. Meanwhile, drought stress response elements such as ABRE and W-box were almost present in the promoter fragments of those significant different expressed NtUGT genes, which suggested their potential roles in drought resistance. In general, combined with the drought response element and the expression patterns of NtUGTs under drought stress, suggested that most of the different expressed NtUGTs might have repressive roles in drought resistance.
Several genes involved in the general flavonoid biosynthesis pathway have been characterized in tobacco. However, enzymes encoded by UGT genes identified as catalyzing the final glycosylation steps in flavonoid biosynthesis were rare, and remained to be fully characterized. Members of the UGT71/72/73/75/78/79/83/91/94 families have been identified in flavonoid biosynthesis pathways in a range of plants [18, 20, 46-49]. In this study, we investigated the involvement of NtUGT genes in flavonoid biosynthesis in two ways. First, we analyzed RNA-Seq data from pink and white tobacco flowers, from which we identified 22 differentially expressed NtUGTs. Among them, except for NtUGT genes included in UGT71 (NtUGT217), UGT73 (NtUGT2), UGT74 (NtUGT123, NtUGT195, and NtUGT224), UGT91 (NtUGT60) family, NtUGT genes in the UGT89 family (NtUGT124, NtUGT127, and NtUGT179) and in the UGT90 family (NtUGT20, NtUGT86, and NtUGT245) were identified for first time as differentially expressed in pink or white flowers. This indicated that they functioned in flavonoid biosynthesis. Second, we performed PPI network analysis, using NtUGTs as bait proteins to identify interacting proteins. NtUGT60, NtUGT94, NtUGT162, NtUGT170, and NtUGT192 were found to interact with some key genes involved in flavonoid biosynthesis, including CHS, CHI, LDX, and three P450 genes. This suggested that those NtUGT genes may function in flavonoid biosynthesis.
Some NtUGT genes were detected as differentially expressed in multiple conditions, namely in response to both cold and drought; in response to drought and between differently-colored flowers; or in response to cold and between differently-colored flowers. Nine NtUGT genes were differentially expressed in response to both cold and drought treatments. Two UGT86A1 genes (NtUGT56 and NtUGT187), NtUGT266, and NtUGT206 were significantly down-regulated whereas NtUGT98 was significantly up-regulated under both conditions. Moreover, all of these five significant different genes contains ABRE motif in their promoter fragment, which a key cis-element response to cold and drought stress. These results suggested that NtUGT56, NtUGT187, NtUGT206, NtUGT266 and NtUGT98 may have an important function in cold and drought resistance. Two NtUGT genes were differentially expressed in response to drought and between differently-colored flowers. One ZOG gene (NtUGT127) was down-regulated both in drought and in white flowers. Another UGT90A1 (NtUGT245) was down-regulated both in drought and in pink flowers. These results suggested that NtUGT127 and NtUGT245 may respond to drought stress through modulation of flavonoid contents. Six NtUGT genes were differentially expressed both in cold conditions and in different flower colors; all six were up-regulated under cold stress, but three (NtUGT72, NtUGT179, and NtUGT195) were up-regulated in pink flowers whereas the other three (NtUGT108, NtUGT124, and NtUGT232) were up-regulated in white flowers. These results were similar to those observed in other plants. AtUGT76E11 increases flavonoid accumulation, which enhances drought stress tolerance [49]. AtUGT79B2 and AtUGT79B3 enhance tolerance to low temperature stress by increasing anthocyanin accumulation [34]. MdUGT83L3 increases anthocyanin accumulation in callus tissues and enhances reactive oxygen species (ROS) clearing in response to salt or cold stress exposure [36]. CsUGT91Q2 can regulate the accumulation of flavonols and scavenge ROS in response to cold treatment [28]. In consideration of the correspondence between flavonoid and stress resistance, NtUGT72, NtUGT179, and NtUGT195 may play roles in flavonoid-related cold stress resistance, further investigation of their function in flavonoid biosynthesis and cold resistance should be carried out.
To validate the functions of NtUGT genes predicted to be involved in flavonoid biosynthesis based on the RNA-Seq data, enzymatic analyses were performed. In vitro enzymatic assays validated that seven NtUGT proteins showed activity toward various flavonoid substrates, and they were exist in group L (NtUGT108, NtUGT123, NtUGT155, NtUGT179, and NtUGT195), F (NtUGT217) and F (NtUGT141). The enzymatic analysis results consistent with the previous report that UGT proteins enriched in subgroups A, F, L and E were related to flavonoid synthesis and polyphenolic metabolism [4, 5]. All seven selected NtUGTs exhibited activity on myricetin; six NtUGTs (all except NtUGT217) showed activity on cyanidin; and three NtUGTs (NtUGT108, NtUGT195, and NtUGT217) were active on flavonol aglycones, namely kaempferol and quercetin. NtUGT108, NtUGT195, and NtUGT217 catalyzed kaempferol and quercetin more efficiently than myricetin. Compared with NtUGT108 and NtUGT195, NtUGT217 was more efficient on kaempferol and quercetin. NtUGT108 was more efficient on kaempferol than quercetin, whereas the reverse was true of NtUGT195 (Table 4 and Fig. 6). The products of quercetin catalysis by NtUGT108 or NtUGT195 were mainly Q4Gs with some Q3Gs, whereas NtUGT217 primarily produced Q3Gs (Fig. 6). Catalysis of kaempferol by NtUGT108, NtUGT195, and NtUGT217 primarily produced K3Gs and rare K7Gs. With respect to regio-specificity, NtUGT217 belonged to the UGT71 family; these function similarly to UGT71 proteins in strawberry, which prefer 3-hydroxycoumarin as substrates and form 3-glucosides in the flavonol pathway [50]. NtUGT108 and NtUGT195 belonged to the UGT74 family, and were similar to several UGT74 proteins in other plants that produced 3-O-glucosides in the kaempferol pathway [51]; however, they produced Q4G in tobacco, which differed from the quercetin pathways in other plants. Furthermore, NtUGT217 overexpression was conducted in tobacco to validate its function in flavonoid biosynthesis in vivo. Transgenic lines overexpressing NtUGT217 showed marked increases in quercetin-3-O-glucoside, quercetin-3-O-rutinoside and kaempferol-3-O-rutinoside accumulation, consistent with the enzymatic activities observed in vitro (Fig. 7).
Conclusion
In this study, 276 UGT genes were identified in N. tabacum and were found to form 18 subfamilies. We characterized the chromosomal distribution and gene structure of the genes, predicted their interaction protein using PPI analysis, and then identified NtUGT genes which involved in cold and drought tolerance and in flavonoid biosynthesis based on the online RNAseq data. We identified seven NtUGTs that potentially involved in flavonoid glycosylation, and we further investigated the enzymatic products and enzymatic properties of three NtUGT proteins (NtUGT108, NtUGT195, and NtUGT217), which mainly formed 3- or 4- glucosides towards flavonols. Finally, we over-expressed NtUGT217 in tobacco to validate its function in flavonol biosynthesis. NtUGT217 overexpression transgenic lines showed increased quercetin-3-O-glucoside, quercetin-3-O-rutinoside and kaempferol-3-O-rutinoside accumulation. This study provides a comprehensive description of NtUGT genes in tobacco and explores the key candidate NtUGT genes likely to be involved in cold and drought resistance and in flavonoid biosynthesis. This study lays the groundwork for future functional investigations of NtUGT genes in N. tabacum.
Methods
Identification of NtUGT genes in Nicotiana tabacum
To identify candidate NtUGT genes in tobacco, the Hidden Markov Model (HMM) profile corresponding to the UDPGT domain (PF00201) was retrieved from Pfam (http://pfam.xfam.org/). The N. tabacum protein database (https://solgenomics.net/ftp/genomes/Nicotiana_tabacum/edwards_et_al_2017/annotation/) was searched using the HMM file and hmmsearch software with an E-value threshold of 1e-5. The conserved PSPG motif sequence (44 aa in length) was also used as a query in BLASTP against the N. tabacum protein database. Candidate proteins yielded by the two strategies were screened using SMART (http://smart.emblheidelberg.de) to remove proteins without a complete PSPG motif. All of the identified NtUGT genes were classified and named based on chromosomal location (or scaffold location as necessary). MapChart was used to plot images of the physical gene locations in N. tabacum. The subcellular localization of each NtUGT protein was predicted with PSORT II Prediction (http//www.genscript.com/psort.html).
Gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) biochemical pathways were analyzed for all putative NtUGT genes in tobacco. GO term enrichment among candidate genes was analyzed using the ‘GOseq’ package [52] in R. KOBAS software was used to find statistically enriched KEGG pathways among the candidate genes [53].
Phylogenetic tree construction
Candidate UGT protein sequences from tobacco and Arabidopsis were aligned with ClustalW in MEGA7 (https://www.megasoftware.net/). 114 Arabidopsis UGT protein sequences were retrieved from CAZy (http://www.cazy.org/GlycosylTransferases.html). Sequences that were too short (< 60 aa) or too divergent from the others were removed from the input file after the initial alignment, then the remaining sequences were re-aligned. Phylogenetic analysis was performed in MEGA7 using the neighbor-joining method with 1000 bootstrap replicates.
Analysis of gene structure and conserved motifs in NtUGT genes
Gene structure data comprised information about exons/introns and untranslated region (UTR) organization. These data were obtained for the predicted NtUGT genes from the N. tabacum GFF annotation file and illustrated using the Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/) [54]. The conserved motifs of putative UGT proteins were predicted using MEME (http://meme-suite.org/tools/meme) with a maximum of 10 motifs per sequence. Cis-acting elements, including promoters, enhancers, regulatory sequences, and inducible elements, were identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), then mapped with the GSDS.
Ka/Ks ratio analysis of NtUGT genes
The ratios of non-synonymous substitution rates (Ka) to synonymous substitution rates (Ks) were calculated for NtUGT gene pairs in Phylogenetic Analysis by Maximum Likelihood (PAML) [55] to estimate the selection modes. Ka/Ks ratios greater than, equal to, and less than 1 were considered to represent positive, neutral, and negative selection, respectively.
Prediction and analysis of NtUGT protein interactors
The Ortho venn tool (http://www.bioinfogenome.net/OrthoVenn/) was used to identify orthologous UGT gene pairs between tobacco and Arabidopsis and to establish a homologous mapping relationship. Protein interaction networks were then built for NtUGTs based on the orthologous genes between tobacco and Arabidopsis using STRING software (http://string-db.org/cgi). Finally, predicted interaction networks were displayed in Cytoscape (https://cytoscape.org/).
Differential expression of NtUGT genes involved in flavonoid biosynthesis and in drought and cold stress responses
Transcriptome data were obtained from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), namely the datasets PRJNA590063 (white and pink tobacco flowers), PRJNA534356 (drought-treated and control tobacco plants) and PRJNA368913 (cold-treated and control tobacco plants). The data were analyzed using DESeq2 software [56] to identify differentially expressed genes. Heatmaps were generated using thresholds of fold-change > 1.5 and p < 0.05.
For quantitative realtime (qRT) PCR analysis, tobacco plants with 4 or 5 fully expanded leaves at 4 weeks were used in cold and drought experiment, leave samples were collected after 24 h-cold-treatment or 48 h-drought treatment, respectively. Corolla of white and pink tobacco flowers were collected, all samples were frozen in liquid nitrogen and stored in -80 ℃ for RNA extraction. qRT-PCR was conducted as described by Chen et al. [57, 58].The Tob103 gene (GenBank accession no. U60495) served as an internal control. Primers for the random selected 9 NtUGT genes were listed in Supplementary Table S1. The comparative cycle threshold (2−ΔΔCT) method was used to calculate relative expression levels of target genes.
Expression and purification of NtUGT proteins in Escherichia coli
Primers were designed for 7 candidate NtUGT genes (NtUGT108, NtUGT123, NtUGT141, NtUGT155, NtUGT179, NtUGT195, and NtUGT217) based on the identified coding region sequences (Supplementary Table S2). The resulting PCR products were purified, then ligated to a pGEX4.0 vector digested with the same restriction enzyme EcoR I. The pGEX4.0-NtUGT vectors were sequenced and transformed into E. coli strain BL21 competent cells. Recombinant glutathione S-transferase (GST) fusion protein expression was induced with 60 μl of 50 mg/ml isopropyl β-d-1-thiogalactopyranoside (IPTG). After overnight incubation at 16 °C with shaking at 180 rpm, cells were harvested by centrifugation at 4 °C, then stored at -80 °C prior to purification.
Proteins were affinity-purified using a GST protein fusion and purification system. In brief, cell pellets were re-suspended in lysis/wash buffer containing 50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 1 mM dithiothreitol (DTT) at pH 7.4. Samples were then sonicated for 30 min with a 3 s interval stop each cycle on ice. Crude protein extracts were loaded into a column packed with GST-binding beads to bind the GST fusion proteins, which were eluted with lysis/wash buffer. The recombinant fusion NtUGT proteins were detected via SDS-PAGE and visualized by staining with Coomassie brilliant blue. Protein concentration was measured using bovine serum albumin (BSA) as the standard.
Enzymatic assay and product identification
Recombinant NtUGT proteins (40 μg each) were incubated at 30 °C with 100 mM Tris–HCl (pH 7.0), 0.5 mM substrate, and 5 mM UDP-glucose in a final volume of 500 µl. After 30 min, reactions were stopped with the addition of 500 μl methanol, then centrifuged at 2200 × g for 10 min. Samples were analyzed using high-performance liquid chromatography (HPLC).
For kinetic analysis, purified 15 μg each of NtUGT108, NtUGT195 and NtUGT217 were added to reaction mixtures with 100 mM Tris–HCl (pH 7.0) and 5 mM UDP-glucose in a final volume of 500 µl. The concentration of the tested flavonoid substrates ranged from 0–400 µM (0, 10, 50, 100, 150, 200, 250, 300, 350, 400 µM). Reactions were stopped with addition of 500 µl methanol after 30 min incubation at 30 °C. Samples were centrifuged at 2200 xg for 10 min and further analyzed by HPLC. The Kinetic parameters Vmax and Km were calculated by using GraphPad Prism.
Enzymatic products were separated via HPLC with a linear A:B elution gradient from 95% solvent A (0.2% acetic acid) to 95% solvent B (100% acetonitrile) over a 48 min period with a flow rate of 1 ml min−1. Reaction products were monitored using a diode array detector at 345 nm for flavonols (quercetin, kaempferol, and myricetin) and at 275 nm for anthocyanin.
Subcellular localization of NtUGT217
To determine the subcellular localization of pRI101-NtUGT217-eGFP, the primer pair NtUGT217-eGFP-F/NtUGT217-eGFP-R (Supplementary Table S3) was used to amplify the NtUGT217 coding region. The PCR product was cloned into the BamHI-digested pRI101-eGFP plasmid using the ClonExpress Entry One Step Cloning Kit (Vazyme Biotech, China). Agrobacterium tumefaciens strain EHA105 was transformed with the pRI101-NtUGT217-eGFP construct; Nicotiana benthamiana leaves were infiltrated with transformed bacteria and examined as described by Chen et al. [58].
NtUGT217 overexpression in tobacco
To construct the overexpression vector p1305-NtUGT217, the primer pair NtUGT217-p1305-F/NtUGT217-p1305-R (Supplementary Table S3) was used to amplify the NtUGT217 coding region. The open reading frame (ORF) region of the NtUGT217 gene driven by the double cauliflower mosaic virus (CaMV) 35S promoter was inserted into the BamHI-digested binary vector pCAMBIA1305. Agrobacterium strain LBA4404 was transformed with the p1305-NtUGT217 construct, then tobacco was transformed using the leaf disc method as reported by Horsch et al. [59].
Measurement of quercetin-3-O-glucoside, quercetin-3-O-rutinoside and kaempferol-3-O-rutinoside via HPLC
Quercetin-3-O-glucoside, quercetin-3-O-rutinoside and kaempferol-3-O were extracted and measured as described by Chen et al. [58]. Analyses were carried out on three independent biological replicates, for which there were three technical replicates each.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request. Formal identification of plant materials was undertaken by the corresponding author of this paper (Shuai Chen). No voucher specimen of this material has been deposited in a publicly available herbarium.
References
Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5(9):380–6. https://doi.org/10.1016/s1360-1385(00)01720-9.
Wu B, Gao L, Gao J, Xu Y, Liu H, Cao X, et al. Genome-wide identification, expression patterns, and functional analysis of UDP glycosyltransferase family in peach (Prunus persica L. Batsch). Front Plant Sci. 2017;22(8):389. https://doi.org/10.3389/fpls.2017.00389.
Yonekura SK, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011;66(1):182–93. https://doi.org/10.1111/j.1365-313X.2011.04493.x.
Li Y, Baldauf S, Lim EK, Bowles DJ. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem. 2001;276(6):4338–43. https://doi.org/10.1074/jbc.M007447200.
Li Y, Li P, Wang Y, Dong R, et al. Genome-wide identification and phylogenetic analysis of family-1 UDP glycosyltransferases in maize (Zea mays). Planta. 2014;239(6):1265–79. https://doi.org/10.1007/s00425-014-2050-1.
Zhang Z, Zhuo X, Yan X, Zhang Q. Comparative genomic and transcriptomic analyses of family-1 UDP glycosyltransferase in Prunus Mume. Int J Mol Sci. 2018;19(11):3382. https://doi.org/10.3390/ijms19113382.
He Y, Ahmad D, Zhang X, Zhang Y, Wu L, Jiang P, et al. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018;18(1):67. https://doi.org/10.1186/s12870-018-1286-5.
Mamoon RH, Amjad NM, Bao L, Hussain SZ, Lee JM, Ahmad MQ, et al. Genome-wide analysis of Family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. Plant Physiol. 2016;206:87–97. https://doi.org/10.1016/j.jplph.2016.08.017.
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, et al. The genome of the domesticated apple (Malus x domestica Borkh). Nat Genet. 2010;42(10):833–9. https://doi.org/10.1038/ng.654.
Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, et al. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 2007;52(6):1154–68. https://doi.org/10.1111/j.1365-313X.2007.03307.x.
Rehman HM, Nawaz MA, Shah ZH, Ludwig-Müller J, Chung G, et al. Comparative genomic and transcriptomic analyses of family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci Rep. 2018;8(1):1875. https://doi.org/10.1038/s41598-018-19535-3.
Rahimi S, Kim J, Mijakovic I, Jung KH, et al. Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol Adv. 2019;37(7):107394. https://doi.org/10.1016/j.biotechadv.2019.04.016.
Yonekura SK, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, et al. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012;9(1):154–67. https://doi.org/10.1111/j.1365-313X.2011.04779.x.
Song C, Zhao S, Hong X, Liu J, Schulenburg K, Schwab WA. UDP-glucosyltransferase functions in both acylphloroglucinol glucoside and anthocyanin biosynthesis in strawberry (Fragaria × ananassa). Plant J. 2016;85(6):730–42. https://doi.org/10.1111/tpj.13140.
Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, et al. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 2011;65(1):106–18. https://doi.org/10.1111/j.1365-313X.2010.04409.x.
Frydman A, Weisshaus O, Bar-Peled M, Huhman DV, et al. Citrus fruit bitter flavors: isolation and functional characterization of the gene Cm1, 2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J. 2004;40(1):88–100. https://doi.org/10.1111/j.1365-313X.2004.02193.x.
Frydman A, Liberman R, Huhman DV, Carmeli-Weissberg M, et al. The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: evolution of branch-forming rhamnosyltransferases under domestication. Plant J. 2013;73(1):166–78. https://doi.org/10.1111/tpj.12030.
Su X, Shen G, Di S, Dixon RA, Pang Y, et al. Characterization of UGT716A1 as a multi-substrate UDP: flavonoid glucosyltransferase gene in Ginkgo biloba. Front Plant Sci. 2017;8:2085. https://doi.org/10.3389/fpls.2017.02085.
Yin Q, Shen G, Di S, Fan C, Chang Z, Pang Y. Genome-wide identification and functional characterization of udp-glucosyltransferase genes involved in flavonoid biosynthesis in Glycine max. Plant Cell Physiol. 2017;58(9):1558–72. https://doi.org/10.1093/pcp/pcx081.
Liu X, Lin C, Ma X, Tan Y, Wang J, Zeng M, et al. Functional characterization of a flavonoid glycosyltransferase in sweet orange (Citrus sinensis). Front Plant Sci. 2018;9:166. https://doi.org/10.3389/fpls.2018.00166.
Chen J, Yuan Z, Zhang H, Li W, Shi M, Peng Z, et al. Cit 1, 2RhaT and two novel CitdGlcTs participate in flavor-related flavonoid metabolism during citrus fruit development. J Exp Bot. 2019;70(10):2759–71. https://doi.org/10.1093/jxb/erz081.
Dong T, Hwang I. Contribution of ABA UDP-glucosyltransferases in coordination of ABA biosynthesis and catabolism for ABA homeostasis. Plant Signal Behav. 2014;9(7):e28888. https://doi.org/10.4161/psb.28888.
Mateo-Bonmatí E, Casanova-Sáez R, Šimura J, Ljung K. Broadening the roles of UDP-glycosyltransferases in auxin homeostasis and plant development. New Phytol. 2021;232(2):642–54. https://doi.org/10.1111/nph.17633.
Khorolragchaa A, Kim YJ, Rahimi S, et al. Grouping and characterization of putative glycosyltransferase genes from Panax ginseng Meyer. Gene. 2014;536(1):186–92. https://doi.org/10.1016/j.gene.2013.07.077.
Ma LL, Shang Y, Cao AZ, Qi ZJ, Xing LP, Chen PD, et al. Molecular cloning and characterization of an up-regulated UDP-glucosyltransferase gene induced by DON from Triticum aestivum L. cv. Wangshuibai. Mol Biol Rep. 2010;37(2):785–95. https://doi.org/10.1007/s11033-009-9606-3.
Schweiger W, Pasquet JC, Nussbaumer T, Paris MP, Wiesenberger G, Macadré C, et al. Functional aracterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Mol Plant Microbe Interact. 2013;26(7):781–92. https://doi.org/10.1094/MPMI-08-12-0205-R.
Zhao M, Jin J, Gao T, Zhang N, Jing T, et al. glucosyltransferase csugt78a14 regulates flavonols accumulation and reactive oxygen species scavenging in response to cold stress in Camellia sinensis. Front Plant Sci. 2019;10:1675. https://doi.org/10.3389/fpls.2019.01675.
Zhao MY, Zhang N, Gao T, Jin JY, Jing TT, Wang JM, et al. Sesquiterpene glucosylation mediated by glucosyltransferase UGT91Q2 is involved in the modulation of cold stress tolerance in tea plants. New Phytol. 2020;226(2):362–72. https://doi.org/10.1111/nph.16364.
Mohnike L, Rekhter D, Huang W, Feussner K, Tian H, et al. The glycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis and plant immunity. Plant Cell. 2021;33(3):735–49. https://doi.org/10.1093/plcell/koaa045.
Kantharaj V, Ramasamy NK, Yoon YE, Cheong MS, Kim YN, Lee KA, Kumar V, et al. Auxin-glucose conjugation protects the rice (Oryza sativa L) Seedlings Against hydroxyurea-induced phytotoxicity by activating UDP-glucosyltransferase enzyme. Front Plant Sci. 2022;12:767044. https://doi.org/10.3389/fpls.2021.767044.
Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012;69(6):1030–42. https://doi.org/10.1111/j.1365-313X.2011.04853.x.
Barvkar VT, Pardeshi VC, Kale SM, Kadoo NY, Gupta VS. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genomics. 2012;13:175. https://doi.org/10.1186/1471-2164-13-175.
Pineau E, Sauveplane V, Grienenberger E, Bassard JE, Beisson F, Pinot F. CYP77B1 a fatty acid epoxygenase specific to flowering plants. Plant Sci. 2021;307:110905. https://doi.org/10.1016/j.plantsci.2021.110905.
He X, Zhao X, Gao L, Shi X, Dai X, Liu Y, Xia T, Wang Y, et al. isolation and characterization of key genes that promote flavonoid accumulation in purple-leaf tea (Camellia sinensis L.). Sci Rep. 2018;8(1):130. https://doi.org/10.1038/s41598-017-18133-z.
Li P, Li YJ, Zhang FJ, Zhang GZ, Jiang XY, Yu HM, Hou BK, et al. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017;89(1):85–103. https://doi.org/10.1111/tpj.13324.
Zhang J, An H, Zhang X, Xu F, Zhou B, et al. Transcriptomic analysis reveals potential gene regulatory networks under cold stress of loquat (Eriobotrya japonica Lindl). Front Plant Sci. 2022;13:944269. https://doi.org/10.3389/fpls.2022.944269.
Li Y, Li P, Zhang L, Shu J, Court MH, Sun Z, Jiang L, Zheng C, Shu H, Ji L, Zhang S, et al. Genome-wide analysis of the apple family 1 glycosyltransferases identified a flavonoid-modifying UGT, MdUGT83L3, which is targeted by MdMYB88 and contributes to stress adaptation. Plant Sci. 2022;321:111314. https://doi.org/10.1016/j.plantsci.2022.111314.
Liu Q, Dong GR, Ma YQ, Zhao SM, Liu X, Li XK, Li YJ, Hou BK. Rice glycosyltransferase gene UGT85E1 Is involved in drought stress tolerance through enhancing abscisic acid response. Front Plant Sci. 2021;12:790195. https://doi.org/10.3389/fpls.2021.790195. eCollection 2021.
Long H, Zheng Z, Zhang Y, Xing P, Wan X, Zheng Y, Li L, et al. An abscisic acid (ABA) homeostasis regulated by its production, catabolism and transport in peanut leaves in response to drought stress. PLoS One. 2019;14(6):0213963. https://doi.org/10.1371/journal.pone.0213963.
Ao B, Han Y, Wang S, Wu F, Zhang J, et al. Genome-wide analysis and profile of UDP-glycosyltransferases family in Alfalfa (Medicago sativa L.) under drought stress. Int J Mol Sci. 2022;23(13):7243. https://doi.org/10.3390/ijms23137243.
Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, et al. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell. 2010;22(8):2660–79. https://doi.org/10.1105/tpc.109.071316.
Liu Z, Yan JP, Li DK, Luo Q, Yan Q, Liu ZB, Ye LM, Wang JM, Li XF, Yang Y, et al. UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2015;167(4):1659–70. https://doi.org/10.1104/pp.15.00053.
Li Q, Yu HM, Meng XF, Lin JS, Li YJ, Hou BK. Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis. Plant Biol (Stuttg). 2018;20(1):10–9. https://doi.org/10.1111/plb.12627.
Li Y, Li P, Wang T, Zhang F, Huang X, Hou B. The maize secondary metabolism glycosyltransferase UFGT2 modifies flavonols and contributes to plant acclimation to abiotic stresses. Ann Botany. 2018;122(7):1203–17. https://doi.org/10.1093/aob/mcy123.
Sun YF, Ji K, Liang B, Du YW, Jiang L, Wang J, Kai WB, et al. Suppressing ABA uridine diphosphate glucosyltransferase (SlUGT75C1) alters fruit ripening and the stress response in tomato. Plant J. 2017;91(4):574–89. https://doi.org/10.1111/tpj.13588.
Wang X, Li C, Zhou Z, Zhang Y, et al. Identification of three (Iso) flavonoid glucosyltransferases from Pueraria lobata. Front Plant Sci. 2019;10:28. https://doi.org/10.3389/fpls.2019.00028.
Speeckaert N, El Jaziri M, Baucher M, Behr M, et al. ugt72, a major glycosyltransferase family for flavonoid and monolignol homeostasis in plants. Biology (Basel). 2022;11(3):441. https://doi.org/10.3390/biology11030441.
Jiang X, Shi Y, Dai X, Zhuang J, Fu Z, Zhao X, Liu Y, Gao L, Xia T, et al. Four flavonoid glycosyltransferases present in tea overexpressed in model plants Arabidopsis thaliana and Nicotiana tabacum for functional identification. J Chromatogr B Analyt Technol Biomed Life Sci. 2018;1100–1101:148–57. https://doi.org/10.1016/j.jchromb.2018.09.033.
Knoch E, Sugawara S, Mori T, Nakabayashi R, Saito K, Yonekura-Sakakibara K, et al. UGT79B31 is responsible for the final modification step of pollen-specific flavonoid biosynthesis in Petunia hybrida. Planta. 2018;247(4):779–90. https://doi.org/10.1007/s00425-017-2822-5.
Song C, Gu L, Liu J, Zhao S, Hong X, Schulenburg K, Schwab W, et al. Functional characterization and substrate promiscuity of UGT71 glycosyltransferases from strawberry (Fragaria × ananassa). Plant Cell Physiol. 2015;56(12):2478–93. https://doi.org/10.1093/pcp/pcv151.
Le DD, Kim W, Lim S, Kim SC, Choi G, et al. Identification of three groups of ginsenoside biosynthetic UDP-glycosyltransferases from Gynostemma pentaphyllum. Plant Sci. 2021;313:111069. https://doi.org/10.1016/j.plantsci.2021.111069.
Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. https://doi.org/10.1186/gb-2010-11-2-r14.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. https://doi.org/10.1093/nar/28.1.27.
Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Hereditas. 2007;29(8):1023–6.
Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550.
Chen S, Li YT, Cui LJ, Zhang YC, Zhang ZM, et al. Identification and characterization of chalcone synthase gene family members in Nicotiana tabacum. J Plant Growth Regul. 2017;36:374–84. https://doi.org/10.3389/fpls.2016.01089.
Chen S, Wu FY, Li YT, Qian YL, Pan XH, et al. NtMYB4 and NtCHS1 are critical factors in the regulation of flavonoid biosynthesis and are involved in salinity responsiveness. Front Plant Sci. 2019;10:178. https://doi.org/10.3389/fpls.2019.00178.
Horsch RF, Fry JE, Hoffffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–31. https://doi.org/10.1126/science.227.4691.1229.
Acknowledgements
We thank Changqing Yang from Tobacco Research Institute of Chinese Academy of Agricultural Sciences for his service and several excellent suggestions.
Funding
This work was supported by The Agricultural Science and Technology Innovation Program (ASTIP-TRIC01), Natural Science Foundation of Shandong Province (ZR2019BC012), Science Foundation for Young Scholars of Tobacco Research Institute of Chinese Academy of Agricultural Sciences (2021B03). China Tobacco Genome Project (110202101001 (JY-01)) and Central Public-interest Scientific Institution Basal Research Fund (1610232022008).
Author information
Authors and Affiliations
Contributions
SC and AY conceived and designed the research. QY, YZ, XQ performed most of the experiments. SC, FW, XL, MR, XW and YT performed the research. SC, QY and YZ analyzed the data and wrote the paper. YC and AY support the research. The author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All study protocols for plant specimens comply with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file1: Table S1.
Primers used for qRT-PCR amplification.
Additional file 2: Table S2.
Primers used for NtUGT genes amplification.
Additional file 3: Table S3.
Primers used in vector construction to determine NtUGTs subcellular localization and for NtUGT217 overexpression.
Additional file 4: Table S4.
Subcellular location of 276 NtUGTs in Nicotiana tabacum.
Additional file 5: Table S5.
Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) biochemical pathways associated with NtUGTs.
Additional file 6: Table S6.
Intron/exon data for NtUGT genes.
Additional file 7: Figure S1.
The predicted subcellular location distribution of NtUGT genes.
Additional file 8: Figure S2.
GO and KEGG analysis of NtUGT genes in Nicotiana tabacum. A. GO term enrichment analysis results. B. KEGG pathway enrichment analysis results.
Additional file 9: Figure S3.
Chromosome distribution of 123 NtUGT genes in Nicotiana tabacum. NtUGTs were distributed across 24 chromosomes. Green colored bars represent chromosomes; chromosome numbers are given at the top of each bar. Red boxes indicate genes derived from tandem duplication.
Additional file 10: Figure S4.
Gene structure analysis of NtUGT genes in Nicotiana tabacum. A.Gene structure and their phylogenetic results of NtUGT genes. Green box indicates exons and dark lines indicates introns of NtUGT genes. B. Conserved motif analysis of NtUGT proteins. Motifs were marked by different colors.
Additional file 11: Figure S5.
Cis-elements distribution of NtUGT genes in Nicotiana tabacum.
Additional file 12: Figure S6.
Relative expression levels of selected NtUGT genes. A. Relative expression levels of selected NtUGT genes in response to cold and drought treatments. B. Relative expression levels of selected NtUGT genes in white and pink tobacco flowers.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, Q., Zhang, Y., Qu, X. et al. Genome-wide analysis of UDP-glycosyltransferases family and identification of UGT genes involved in abiotic stress and flavonol biosynthesis in Nicotiana tabacum. BMC Plant Biol 23, 204 (2023). https://doi.org/10.1186/s12870-023-04208-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04208-9