Abstract
Background
Flavonoids are secondary metabolites that play significant roles in plant cells. In particular, polymethoxy flavonoids (PMFs), including nobiletin, have been reported to exhibit various health-supporting properties such as anticancer, anti-inflammatory, and anti-pathogenic properties. However, it is difficult to utilize PMFs for medicinal and dietary use because plant cells contain small amounts of these compounds. Biosynthesis of PMFs in plant cells is carried out by the methylation of hydroxyl groups of flavonoids by O-methyltransferases (FOMT), and many kinds of FOMTs with different levels of substrate specificity and regioselectivity are cooperatively involved in this biosynthesis.
Results
In this study, we isolated five genes encoding FOMT (CdFOMT1, 3, 4, 5, and 6) from Citrus depressa, which is known to accumulate nobiletin in the peels of its fruits. The genes encoded Mg2+-independent O-methyltransferases and showed high amino acid sequence similarity (60–95 %) with higher plant flavonoid O-methyltransferases. One of these genes is CdFOMT5, which was successfully expressed as a soluble homodimer enzyme in Escherichia coli. The molecular mass of the recombinant CdFOMT5 subunit was 42.0 kDa including a 6× histidine tag. The enzyme exhibited O-methyltransferase activity for quercetin, naringenin, (-)-epicatechin, and equol using S-adenosyl-l-methionine (SAM) as a methyl donor, and its optimal pH and temperature were pH 7.0 and 45 °C, respectively. The recombinant CdFOMT5 demonstrated methylation activity for the 3-, 5-, 6-, and 7-hydroxyl groups of flavones, and 3,3′,5,7-tetra-O-methylated quercetin was synthesized from quercetin as a final product of the whole cell reaction system. Thus, CdFOMT5 is a O-methyltransferase possessing a broad range of substrate specificity and regioselectivity for flavonoids.
Conclusions
Five FOMT genes were isolated from C. depressa, and their nucleotide sequences were determined. CdFOMT5 was successfully expressed in E. coli cells, and the enzymatic properties of the recombinant protein were characterized. Recombinant CdFOMT5 indicated O-methyltransferase activity for many flavonoids and a broad regioselectivity for quercetin as a substrate. Whole-cell biocatalysis using CdFOMT5 expressed in E. coli cells was performed using quercetin as a substrate, and 3,3′,5,7-tetramethylated quercetin was obtained as the final product.
Similar content being viewed by others
Background
Flavonoids are major secondary metabolites in plants. More than 10,000 kinds of flavonoid derivatives are estimated to occur in plants [1]. Such varieties of flavonoid subgroups include flavones, flavonols, flavanones, flavanes, flavanols, isoflavones, and anthocyanidins, all of which originate from the phenylpropanoid synthesis pathway [2, 3]. In plant cells, flavonoids are modified by many enzymes, such as methyltransferases, glycosyltransferases, sulfotransferases, acyltransferases, oxidases, and others. These modifications, coupled with the structural variation observed in different flavonoid subgroups, contribute to a huge diversity of flavonoids [4, 5].
It is well known that many flavonoids have significant roles in plants, such as inflorescence pigments and antioxidants as well as serving as substances with anti-pathogenic [6, 7], anti-insect [8], and signaling functions [9]. Recently, it has been reported that O-methylated flavonoids have significant biological activity in humans, exhibiting antibiotic [10], antiviral [11], anti-cancer [12, 13], anti-inflammatory [14], anti-obesity [15, 16], neuroprotective [17, 18], and anti-allergy [19] properties. Increasing evidence of the effects of O-methylated flavonoids on human health suggests they can be used to enrich processed foods or in dietary supplements and pharmaceuticals. Nobiletin (3′,4′,5,6,7,8-hexamethoxyflavone) is one of the abundant polymethoxy flavonoids (PMF) in Citrus depressa peels, and it has been reported to possess several bioactivities [13, 14, 20, 21]. However, it is difficult to investigate nobiletin in general and medical research because of the small amounts of nobiletin obtained from Citrus peels. In plant cells, hydroxyl groups of flavonoids are methylated by reactions with O-methyltransferase (OMT) using S-adenosyl-l-methionine (SAM) as a methyl donor.
Plant OMTs are generally classified as either class I or class II by their structural and enzymatic properties [22]. Class I OMTs, which include the well-known enzyme caffeoyl-CoA 3-OMT (CCoAOMT), are characterized by low subunit molecular masses (from 23 to 27 kDa), dependence on Mg2+ ions, and the ability to catalyze the methylation of 3-hydroxyl groups of caffeoyl-CoA to produce feruloyl-CoA. Thus, CCoAOMT, which is involved in lignin biosynthesis in plant cells, is the key enzyme for producing monolignols [23, 24]. Class II OMTs have a higher subunit molecular mass than CCoAOMTs (from 38 to 43 kDa) and do not require Mg2+ ions for methylation. Plant cells accumulate many flavonoids through the reactions of such OMTs. Ibrahim et al. reported that cell-free extracts of Citrus mitis exhibited stepwise O-methylation activities for various flavonoids and suggested that different types of OMTs are involved in the biosynthesis of PMFs in some tissues (peel, root, and callus tissue) [25, 26]. More recently, it was reported that there are 58 OMT genes in the C. sinensis (sweet orange) genome, and these genes show distinct expression patterns that differ among tissues and developmental stages [27]. These findings strongly suggested that the structural diversity of PMFs in citrus is caused by combinations of various types of substrate- and regio-specific methyltransferases.
Here, we report the isolation and characterization of five flavonoid O-methyltransferase (FOMT) genes (CdFOMT1, 3, 4, 5, and 6) from C. depressa. Furthermore, CdFOMT5 was successfully expressed in E. coli as a functional enzyme, and its properties were characterized in detail. CdFOMT5 possessed methyltransferase activity for quercetin, a ubiquitous flavonoid in plants, and exhibited a broad range of substrate specificity and regioselectivity toward 3-, 5-, 6-, and 7-hydroxyl groups of flavones. Using the biotransformation of quercetin in a CdFOMT5-expressing E. coli biocatalyst, we successfully obtained 3,3′,5,7-tetra-O-methylated quercetin as a final product, suggesting that the enzyme participates in the biosynthesis of nobiletin.
Results and discussion
Cloning of CdFOMT genes from C. depressa
To isolate FOMT-coding genes from C. depressa, we performed degenerate PCR using primers designed from the conserved amino acid sequences of higher plant FOMTs. Using genomic DNA as the template, we obtained five fragments, whose deduced amino acid sequences showed similarity to several higher plant FOMTs. In order to obtain the full-length FOMT genes from C. depressa, we performed thermal asymmetric interlaced PCR (TAIL-PCR) and obtained five FOMT-coding genes, CdFOMT1, 3, 4, 5 and 6. All primers used to clone CdFOMT genes are listed in Additional file 1: Table S1. Then, we amplified full-length cDNA of these CdFOMT genes using specific primers designed from the deduced N-terminal and C-terminal sequences. Thus, we isolated five FOMT genes which encode homologous proteins CdFOMT1, 3, 4, 5, and 6. The amino acid sequences of CdFOMT3, 5, and 6 showed relatively high identities each other (from 67.8 to 82.3 %), although those of CdFOMT1 and 4 showed lower identities (from 28.9 to 39.9 %) with other CdFOMTs (Table 1).
A comparison of the genomic and cDNA sequences revealed that these FOMT genes contain one or three introns (Additional file 2: Figure S1). All CdFOMT genes contain an intron at the same position within their coding sequences [Additional file 2: Figure S1, marked with an asterisk; AIXXK-(intron)-(S/W)(I/V)LHDW], although these CdFOMT genes show quite low similarities across their whole nucleotide sequences, and the proteins differed substantially in size. Schroder et al. [28] found that COMT genes from Catharanthus roseus share an intron insertion location. Similarly, the amino acid sequences at this site in CdFOMT genes were highly conserved across many plant OMTs. CdFOMT3 and CdFOMT5 each contained three introns, which also share positions within their amino acid sequences [QDKXXLXS-(intron1)-WSXLK ~ PHVIXHXPXXP-(intron2)-XXXHVGGDM ~ DAIXXK-(intron3)-(W/S)(I/V)XLHDW], though there were again large differences in both their sizes and sequences as proteins. This result suggests that these CdFOMT genes have the same origin and acquired their introns during their shared evolutionary history.
The five newly isolated CdFOMT genes from C. depressa encode 335 to 362 amino acid residues and shared 60–97 % amino acid sequence identities to known higher plant OMTs (Table 2). Figure 1 shows the alignment of the deduced CdFOMT amino acid sequences and higher plant FOMTs. CdFOMTs exhibited several conserved sequences (motifs A, B, C, J, K, and L in Fig. 1), which are likely involved in interactions with the cofactor SAM [22, 29]. The existence of these conserved regions suggests that the five OMT genes obtained from C. depressa code for potential SAM-dependent FOMTs.
Multiple sequence alignment of CdFOMT sequences with higher plant flavonoid O-methyl transferases (FOMTs): C. depressa (CdFOMT1, DDBJ accession number LC126056; CdFOMT3, LC126057; CdFOMT4, LC126058; CdFOMT5, LC126059; and CdFOMT6, LC126060), Citrus sinensis (CsFOMT, GenBank accession number ABP94018), Arabidopsis thaliana (At3OMT, AED96460), Chrysosplenium americanum (Ca3′OMT1, AAA80579), Mentha × piperita (Mp3′OMT, AAR09601), and Triticum aestivum (Ta3′4′5′OMT, ABB03907). Amino acid sequences were aligned using ClustalW. Black highlighting denotes amino acids that are conserved across many FOMTs
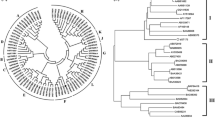
Phylogenetic analysis of five CdFOMTs with 25 putative and defined plant class II OMTs from higher plants (Fig. 2) indicate that they are plant class II OMTs (divalent cation independent). Class II OMTs are generally known to show activity with flavonoids and isoflavonoids [30], while class I OMTs catalyze methylation of phenolic compounds involved in lignin synthesis [23, 24]. These results suggest that the isolated CdFOMTs may be involved in the biosynthesis of PMFs such as nobiletin in C. depressa.
A phylogenetic tree constructed from plant FOMT amino acid sequences. In addition to sequences of CdFOMTs characterized in this paper, 25 plant class II FOMT sequences were also selected using BLAST search and aligned by ClustalW. The phylogenetic tree was constructed using the NJplot program. Arabidopsis thaliana (At3OMT, GenBank accession number AED96460 and AtOMT1, AAB9679), Catharanthus roseus (CrFOMT, AAM97497 and CrOMT6, AAR02419), Chrysosplenium americanum (Ca3′OMT1, AAA80579), Citrus sinensis (CsFOMT, ABP94018), Glycyrrhiza echinata (Ge4′OMT, AB091684 and Ge7IOMT, AB091685), Hordeum vulgare (HvFI7OMT, CAA54616), Lotus japonicus (Lj4′OMT, AB091686), Medicago sativa (Ms3′OMT, AAB46623 and Ms7IOMT, MSU97125), Mentha × piperita (Mp3′OMT, AAR09601; Mp4′OMT, AAR09602; Mp7OMT1A, AAR09598; Mp7OMT1B, AAR09599; and Mp8OMT, AAR09600), Oryza sativa (Os7OMTlike1, BAD29452; Os7OMTlike2, BAD05699; and Os3OMT, BAF22945), Populus trichocarpa (PtFOMT2, EEE86889; PtOMT5, EPR47487; and PtFOMT10, EPR54183), Triticum aestivum (Ta3′4′5′OMT, ABB03907), and Zea mays (ZmCOMT, ABQ58826)
Expression of recombinant CdFOMTs genes in Escherichia coli
In order to obtain recombinant enzymes, each CdFOMT gene was cloned into a pET21b vector, and E. coli BL21(DE3) cells were transformed with a constructed plasmid. With the exception of the CdFOMT5, the four remaining CdFOMTs formed inclusion bodies under several culture conditions and no activity could be detected for these proteins. Schroder et al. [28] reported that three OMT genes in Catharanthus roseus (CrOMT5, 6, and 7) were tested for expression in E. coli, and recombinant CrOMT6 and CrOMT7 formed soluble proteins whereas CrOMT5 formed an insoluble protein. In general, expression of functional plant enzymes in E. coli is unpredictable and must be determined empirically with many factors such as amino acid composition, protein folding, and post translational modifications influencing the outcome. Thus, the newly isolated methyltransferase genes other than CdFOMT5 may contribute to biosynthesis of PMF in C. depressa, but details of their biochemical functions remain unclear. We were able to successfully express recombinant CdFOMT5 in E. coli and confirm its OMT activity. Recombinant CdFOMT5 was obtained as a fusion protein with a 6× histidine tag at the C-terminus and purified by Ni-Sepharose resin column chromatography. Recombinant CdFOMT5 was purified to homogeneity, and a single 42.0-kDa protein band was obtained by an SDS-PAGE analysis (Fig. 3).
Physicochemical properties of CdFOMT5
Using HPLC, the molecular mass of recombinant CdFOMT5 was estimated to be 88.0 kDa. The theoretical molecular mass of the recombinant CdFOMT5 including the 6× histidine tag is 42.03 kDa, which agrees with the observed molecular mass of 42.0 kDa inferred using SDS-PAGE. It is generally known that typical plant FOMT subunits are homodimers [31, 32]. These findings suggest that recombinant CdFOMT5 is a homodimer protein in E. coli cells.
The pI value of the enzyme without the 6× histidine tag based on its amino acid sequence was theoretically calculated to be 5.79.
The effects of pH and temperature on OMT activity were measured using quercetin as a substrate (Additional file 2: Figure S2A). Recombinant CdFOMT5 showed optimum activity at pH 7.0 (in potassium phosphate buffer), and its activity fell more than 50 % at pH 5.5 (in Na-citrate buffer) or pH 9.0 (in Tris-HCl buffer; Additional file 2: Figure S2A). The optimum temperature of CdFOMT5 was 45 °C, and the enzyme exhibited more than 80 % of maximum activity at 55 °C (Additional file 2: Figure S2B).
Regioselectivity of CdFOMT5 for quercetin
When O-methyltransferase activity was measured using quercetin as a substrate, several peaks corresponding to O-methylated products were detected by HPLC analysis (Fig. 4). The retention time for P1 was consistent with 3-O-methylquercetin, and that of P2 was consistent with azaleatin (5-O-methylquercetin) or rhamnetin (7-O-methylquercetin). However, P3 and P4 had retention times that differed from those of quercetin mono-methylated derivatives. To investigate the molecular mass of these compounds, we performed an LC-MS analysis and observed increases of 28- and 42-Da in the molecular ion peaks for P3 and P4, respectively (Additional file 2: Figure S3). This demonstrates that CdFOMT5 can catalyze the O-methylation of at least three hydroxyl groups of quercetin and that di- or tri-O-methylated quercetin products were obtained by this enzymatic reaction.
HPLC analysis of quercetin products methylated by CdFOMT5: S, quercetin; P1, 3-O-methylated quercetin; P2, azaleatin/rhamnetin; P3, dimethylated quercetin; and P4, trimethylated quercetin. The peak labeled with ‘x’ was not identified as corresponding to flavonoid derivatives. (See Additional file 2: Figure S2)
To confirm the regioselectivity of CdFOMT5, the enzymatic reaction was measured using mono-hydroxyflavones as a substrate. As shown in Fig. 5, CdFOMT5 demonstrated methylation activity for 3-, 5-, 6-, and 7-hydroxy groups of flavone. Under the standard assay condition (100 μM substrate), the highest activity was observed for 3-hydroxyflavone (flavonol) based on the peak area of the product, followed by 7-hydroxyflavone with a relative activity of 15.6 % of that of 3-hydroxyflavone and 5-hydroxyflavone with that of 13.5 %. Very weak activity was observed for 6-hydroxyflavone (2 % in comparison with the activity of 3-hydroxyflavone), and no activity was detected for 3′- or 4′-hydroxyflavone and 7-methoxy-8-hydroxyflavone (data not shown). This appears to be the first report of an OMT with the ability to catalyze the O-methylation of four positions of hydroxyl groups in the A and C rings of flavonoids. However, there are some reports of sequential methylation of flavonoids by OMTs [33, 34]. In particular, TaOMT2 from wheat exhibited definite sequential methylation of tricetin at its B-ring 3′-, 5′-, and 4′-hydroxyl groups. However, in the CdFOMT5 reaction, not only 3-O-methylated quercetin but also azaleatin (5-O-methylated quercetin)/rhamnetin (7-O-methylated quercetin) were obtained from quercetin, and this result indicates that CdFOMT5 does not exhibit a sequential methylation order for 3-, 5-, and 7-hydroxyl groups of quercetin.
Nobiletin is a major PMF in C. depressa peel, and its 3′-, 4′-, 5-, 6-, 7-, and 8-hydroxy groups are all methylated. Therefore, CdFOMT5 is hypothesized to play a significant role in the biosynthesis of PMF in C. depressa. There are many reports that a combination of several OMTs catalyze the O-methylation of flavonoids to produce PMFs [35, 36]. Furthermore, Joe et al. [37] reported that mutant OMT (POMT7-M1) exhibits methylation activity for 3- and 7-hydroxy groups of flavonoids, whereas native POMT7 can methylate only the 7-hydroxy group [37]. They also showed that substitution of Asp with Gly at amino acid position 257 significantly affected the regioselectivity of POMT7. Although CdFOMT5 contains Asp260 corresponding to the Asp257 of native POMT7, CdFOMT5 was able to catalyze the methylation reaction of flavonoids more widely than the mutated POMT7. Changes at a few amino acid residues of class II O-methyltransferase also have substantial effects on substrate specificity, kinetic property, and regioselectivity [35, 38–40]. These findings indicate that size or shape of the catalytic center of CdFOMT5 plays an important role in determining the broad regioselectivity of CdFOMT5.
Substrate spectrum of CdFOMT5
To investigate the substrate specificity of CdFOMT5, O-methyltransferase activities for naringenin, (-)-epicatechin, equol, and cyanidin were measured (Additional file 2: Figure S4). Recombinant CdFOMT5 exhibited OMT activity for each of these substrates except cyanidin, although detailed structural determinations of the products have not yet been performed. However, we were not able to detect polymethylated products from naringenin, (-)-epicatechin, and equol, even though CdFMOT5 exhibited a broad regioselectivity toward quercetin and monohydroxylated flavone (Fig. 5). The results suggested that CdFMOT5 prefers flavonol (3-hydroxyflavone) to other flavonoid structures. Thus, substrate structure, especially the C-ring in flavonoids, may strongly affect the substrate preference, including regioselectivity, of CdFOMT5.
Bioproduction of polymethylated quercetin with recombinant E. coli cells expressing CdFOMT5
To examine the production of PMF by an E. coli biocatalyst expressing CdFOMT5, bioconversion was performed using quercetin as a substrate. In the presence of l-methionine and glucose for regenerating SAM in the reaction mixture, many peaks corresponding to polymethylated quercetins were observed. This agreed with the results of enzymatic reactions of mono-hydroxyflavones. In contrast, negligible product amounts were obtained in the absence of methionine and glucose (data not shown). This result clearly indicates that whole recombinant E. coli cells efficiently regenerated SAM using l-methionine and glucose and that the methylation reaction was successfully carried out. Additional peaks that were not expected in the enzymatic reaction were detected (peaks 6 and 7 in Fig. 6a). LC-MS analysis showed that these extra products correspond to tri- and tetra-methylated quercetin (peaks 6 and 7, respectively, in Fig. 6b). Figure 5 shows that CdFOMT5 catalyzes the methylation of the 3-, 5-, and 7-hydroxy groups of monohydroxylated flavones, although it does not catalyzes the 3′ or 4′-monohydroxyflavone (data not shown). Additionally, isorhamnetin (3′-O-methylated quercetin) was a product of E. coli host cells that lacked CdFOMT5 when used for bioconversion as a control (peak 2 in Fig. 6a). Therefore, we surmised that an unexpected 3′-O-methylation reaction occurred in E. coli host cells. Thus, the product corresponding to peak 7 was hypothesized to be 3,3′,5,7-tetramethylated quercetin. To verify this, the product of peak 7 was purified and analyzed by H1-NMR. As shown in Fig. 6c, four singlet peaks corresponding to the four methoxy groups were detected within a concentration range of 3.8 to 4.0 ppm. This strongly suggests that the product corresponding to peak 7 is 3,3′,5,7-tetramethylated quercetin (Fig. 6d).
Bioproduction of polymethylated quercetin using recombinant E. coli carrying CdFOMT5. a The HPLC analysis of the reaction mixture identified five peaks: (1) 3-O-methylated quercetin; (2) isorhamnetin (3′-O-methylated quercetin); (3) azaleatin/tamarixetin; (4) dimethylated quercetin; (5 and 6) trimethylated quercetin; and (7) 3,5,7,3′-tetramethylated quercetin. Peaks labeled with ‘x’ were not identified as quercetin derivatives. Transformant cells harboring empty vector (pET21b) was used as host cell. b Mass spectrometry analysis of purified peak 7 product. c NMR spectrum of purified product (peak 7). d Predicted structure of 3,3′,5,7-O-methylated quercetin
Conclusions
In this study, we successfully obtained five FOMT genes from C. depressa and expressed the CdFOMT5 gene in E. coli cells. Recombinant CdFOMT5 demonstrated SAM-dependent O-methyltransferase activity for quercetin, and its optimum pH and temperature were 7.0 and 45 °C, respectively. CdFOMT5 exhibited a broad range of not only substrate specificity, but also regioselectivity and catalyzed the methylation of 3-, 5-, 6-, and 7-hydroxyl groups of flavone. Furthermore, quercetin was converted to 3,3′,5,7-tetramethylated quercetin as a sequentially methylated product by the E. coli whole cell reaction system. Thus, CdFOMT5 is a useful O-methyltransferase possessing a wide range of regioselectivity for flavonoids and likely plays a role in the synthesis of nobiletin in C. depressa. Further improvement of the host cell through metabolic engineering and the use of engineered CdFOMT5 would make this bioprocess suitable for producing various PMFs.
Methods
Strains and vectors
Fruits of C. depressa Hayata (known as shequasar or Taiwan tangerine) were purchased from an agricultural cooperative in Okinawa, Japan to obtain genomic DNA and total RNA. E. coli JM109 cells and the cloning vectors pGEM-T Easy (Promega, Madison, WI, USA) and pUC118 were used to clone the CdFOMT coding genes. The expression vector pET21b(+) and E. coli BL21(DE3) cells were used to express recombinant CdFOMT genes.
Preparation of genomic DNA and total RNA
Standard techniques were used for DNA manipulation [41]. Genomic DNA and total RNA were obtained from the pericarp of C. depressa fruits. Flesh pericarp was ground using a mortar and pestle in the presence of liquid nitrogen. Genomic DNA was extracted by the cetyltrimethylammonium bromide (CTAB) extraction method [42]. Total RNA was prepared using a TRI reagent (Cosmo Bio Co. Ltd., Tokyo, Japan) according to the manufacturer’s protocol. First-strand cDNA was synthesized using a PrimeScript High Fidelity RT-PCR Kit (TaKaRa, Shiga, Japan) with an oligo dT primer, and the products were used as PCR templates to isolate CdFOMT-coding genes.
Cloning of the CdFOMT-coding genes
To obtain partial fragments of CdFOMT-coding genes, PCR was carried out using genomic DNA as a template. The degenerate primers for amplifying CdFOMT fragments were designed from the amino acid sequences of caffeic acid/flavonoid O-methyltransferases that are conserved among higher plants. To obtain the full-length CdFOMT-coding genes, TAIL-PCR [43] was carried out based on the deduced partial nucleotide sequences. The full lengths of the cDNA fragment of CdFOMT were amplified by PCR using the first-strand cDNA as a template. Amplified fragments were cloned into the pGEM-T Easy vector. All oligonucleotide primers used to clone CdFOMT-coding genes are shown in Additional file 1: Table S1, and all nucleotide sequences were determined using a Capillary DNA Sequencer 3130 (Applied Biosystems, Tokyo, Japan) to perform Sanger DNA sequencing for both strands.
Nucleotide sequence accession numbers
The nucleotide sequences of the isolated CdFOMTs were submitted to the DNA Data Bank of Japan (DDBJ) under the following accession numbers: CdFOMT1, LC126056; CdFOMT3, LC126057; CdFOMT4, LC126058; CdFOMT5, LC126059; and CdFOMT6, LC126060.
Expression and purification of recombinant CdFOMT5
Cloned cDNA fragments of CdFOMT-coding genes were excised by Bam HI and Sal I and purified by agarose gel electrophoresis. Purified DNA fragments were cloned into the expression vector pET21b(+) that had been digested with the same restriction endonucleases. The resulting plasmids were named pET-CdFOMT1 through pET-CdFOMT6, and each of them was introduced into E. coli BL21(DE3) cells.
The transformants harboring the CdFOMT5 gene were grown on LB medium (containing 50 μg/mL ampicillin) to OD660 0.5 at 30 °C with shaking. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM, and the cells were incubated for another 24 h at 18 °C to induce the expression of recombinant CdFOMT5. After induction, cells were collected by centrifugation at 33,800 × g for 5 min and washed with 50 mM potassium phosphate buffer (KPB; pH 7.0). Cells were harvested by centrifugation and resuspended in cell lysis buffer (50 mM KPB, 200 mM NaCl, 10 % glycerol, 10 mM imidazole, pH 7.2). Disruption of cells was carried out by sonication for 30 min at 4 °C using an Insonator 201 M (Kubota, Tokyo, Japan), and cell debris was centrifuged two times at 33,800 × g for 20 min to obtain a clear lysate. The recovered supernatant was applied to Ni-Sepharose chromatography columns (GE Healthcare, Tokyo, Japan; 10 mL bed volume) and washed with 50 mL of the same buffer. Recombinant proteins fused with 6× histidine tags were eluted by the same buffer containing 250 mM imidazole. Collected recombinant CdFOMT5 protein was passed through a desalting column (Econo-Pac PD-10; Bio-Rad Lab., Tokyo, Japan) using 50 mM KPB containing 10 % glycerol. The concentration of the purified recombinant CdFOMT5 was determined using the Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Hercules, CA, USA), and recombinant protein was used for further experiments.
Determination of the molecular mass of recombinant CdFOMT5
To investigate the molecular mass of native recombinant CdFOMT5, an HPLC analysis was carried out using the Shimadzu LC-10 HPLC system (Shimadzu, Kyoto, Japan) equipped with a Superdex 200 10/300 GL Column (GE Healthcare, Port Washington, NY, USA). First, 100 μL of purified recombinant CdFOMT5 solution was applied and separated using the mobile phase [100 mM KPB containing 200 mM NaCl (pH 7.0), 0.4 mL/min]. Protein absorbance was then monitored at 280 nm, and molecular mass was estimated from the retention times of authentic molecular weight markers (Oriental Yeast Co., Ltd., Tokyo, Japan).
Enzyme assay
To determine the flavonoid O-methyltransferase activity of recombinant CdFOMT5, the purified enzyme was tested for its reaction with quercetin, several hydroxyflavones, and some flavonoids in the presence of SAM as a methyl donor. The reaction mixture consisted of 50 mM KPB (pH 7.0), 500 μM SAM, 100 μM substrate (10 mM in dimethylformamide), and 50 μL of purified CdFOMT5, yielding a total volume of 500 μL. The reaction mixture was incubated at 30 °C for 1 h with shaking (at 1,000 rpm), and the reaction was stopped by addition of 500 μL of methanol. After vigorous mixing, the supernatant was recovered by centrifugation and analyzed by HPLC or LC-mass spectrometry (MS). All enzyme assays were performed in triplicate.
Bioproduction of polymethylated quercetin by resting-cell reactions
The E. coli transformant cells expressing recombinant CdFOMT5 were prepared according to the methods described above. Collected cells from 100-mL cultures were washed and resuspended in a M9 salt medium supplemented with 1 % glucose and 10 mM l-methionine at a final cell concentration of 1 % (w/v) in a total volume of 0.5 mL. Then, 100 μM quercetin was added to the reaction mixture as a substrate, and the reaction was carried out at 30 °C for appropriate time with shaking. Reaction products were extracted twice with an equal volume of ethyl acetate and dried over anhydrous sodium sulfate, and then the organic solvent was evaporated in a vacuum. Obtained products were dissolved in methanol and analyzed by HPLC or LC-MS.
HPLC analysis
HPLC analysis of the reaction products was carried out with a Shimadzu LC-10 HPLC system equipped with a Cadenza CD-C18 Column (4.6 mm × 75 mm; Imtakt Corp., Kyoto, Japan). Analytical conditions for quercetin, hydroxyflavones, naringenin, and equol were as follows: eluent A, 0.05 % (v/v) formic acid; eluent B, acetonitrile; flow rate, 0.5 mL/min; column temperature, 40 °C; sample injection, 5 μL; and a linear gradient program (% eluent B) of 0 to 10 min (15 %), 10 to 25 min (10 to 55 %), 25 to 40 min (55 to 75 %), 40 to 45 min (75 to 15 %), and 45 to 55 min (15 %). The absorbances of products were monitored at 254 nm. The retention time of each compound was as follows: 24.8 min for quercetin; 25.8 min for 3-O-methylquercetin; 27.7 min for isorhamnetin (3′-O-methylquercetin); 27.8 min for tamarixetin (4′-O-methylquercetin); 29.7 min for rhamnetin (7-O-methylquercetin)/azaleatin (5-O-methylquercetin); 35.2 min for 3-hydroxyflavone; 34.9 min for 3-methoxyflavone; 37.7 min for 5-hydroxyflavone; 30.7 min for 5-methoxyflavone; 29.3 min for 6-hydroxyflavone; 34.3 min for 6-methoxyflavone; 28.2 min for 7-hydroxyflavone; 33.2 min for 7-methoxyflavone; 26.9 min for naringenin; and 27.1 min for equol. The gradient program (% eluent B) for (-)-epicatechin was as follows: 0 to 5 min (10 %); 5 to 22 min (10 to 25 %); 22 to 27 min (25 %); 27 to 30 min (25 to 10 %); and 30 to 35 min (10 %). The retention time of (-)-epicatechin was 12.5 min. The gradient program (% eluent B) for cyanidin was as follows: 0 to 30 min (5 to 100 %); 30 to 35 min (100 %); 35 to 40 min (100 to 5 %); and 40 to 45 min (5 %). The retention time of cyanidin was 8.6 min. Eluents for analysis of (-)-epicatechin and cyanidin were the same as that used for quercetin.
Mass spectrometry
LC-MS analysis of reaction products was performed using an Agilent 1200 HPLC system (Agilent Technologies, Santa Clara, CA, USA) connected to a Bruker microTOF Mass Spectrometer (Bruker Corporation, Billerica, MA, USA) equipped with an electrospray ionization (ESI) source. The sample (5 μL) was dissolved in methanol and injected into a Cadenza CD-C18 column. Products were separated and eluted under the same HPLC conditions described in HPLC analysis, and positive ion mass spectra were acquired from 50 to 500 m/z.
NMR analysis of polymethylated quercetin
1H-NMR spectra of tetra-O-methylated quercetin were recorded on an AV400 spectrometer (Bruker, Rheinstetten, Germany) in DMSO-d 6 (D, 99.9 % with 0.05 % TMS). All signals are expressed as ppm with TMS as the reference. The 1H-NMR (400 MHz, DMSO-d 6) spectra of 3,3′,5,7-tetra-O-methylated quercetin were as follows;: δ 3.83 (s, 3H), 3.90 (s, 3H), 3.92 (s, 3H), 3.93 (s, 3H), 6.36 (d, J = 2.4 Hz, 1H), 6.66 (d, J = 2.0 Hz, 1H), 7.13 (d, J = 8.8 Hz, 1H), 7.74 (d, J = 2.0 Hz, 1H), 7.79 (dd, J = 2.2, 8.6 Hz, 1H).
Chemicals
SAM and 3-O-methylated quercetin were purchased from Sigma (St. Louis, MO, USA). All other chemicals were purchased from Wako Pure Chemicals Industries (Osaka, Japan).
Abbreviations
- FOMT:
-
Flavonoid O-methyltransferase
- OMT:
-
O-methyltransferase
- PMF:
-
Polymethoxy flavonoid
- SAH:
-
S-adenosyl-l-homocysteine
- SAM:
-
S-adenosyl-l-methionine
References
Tahara S. A Journey of Twenty-Five Years through the Ecological Biochemistry of Flavonoids. Biosci Biotechnol Biochem. 2007;71(6):1387–404.
Koes RE, Quattrocchio F, Mol JNM. The flavonoid biosynthetic pathway in plants: Function and evolution. Bio Essays. 1994;16(2):123–32.
Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol. 2002;5(3):218–23.
Cook NC, Samman S. Flavonoids-Chemistry, metabolism, cardioprotective effects, and dietary sources. J Nutr Biochem. 1996;7(2):66–76.
Peterson JMS, Dwyer J, Dsc RD. Flavonoids: Dietary occurrence and biochemical activity. Nutr Res. 1998;18(12):1995–2018.
Iwashina T. Flavonoid function and activity to plants and other organisms. Biol Sci Space. 2003;17(1):24–44.
Treutter D. Significance of flavonoids in plant resistance: a review. Environ Chem Lett. 2006;4:147–57.
Napal GND, Aarpinella MC, Palacios SM. Antifeedant activity of ethanolic extract from Flourensia oolepis and isolation of pinocembrin as its active principle compound. Bioresour Technol. 2009;100(14):36693673.
Taylor LP, Grotewold E. Flavonoids as developmental regulators. Curr Opin Plant Biol. 2005;8(3):317–23.
Ustün O, Ozçelik B, Akyön Y, Abbasoglu U, Yesilada E. Flavonoids with anti-Helicobacter pylori activity from Cistus laurifolius leaves. J Ethnopharmacol. 2006;108(3):457–61.
Castrillo J, Berghe DV, Carrasco L. 3-methylquercetin is a potent and selective inhibitor of poliovirus RNA synthesis. Virology. 1986;152(1):219–27.
Sato T, Koike L, Miyata Y, Hirata M, Mimaki Y, Sashida Y, Yano M, Ito A. Inhibition of Activator Protein-1 Binding Activity and Phosphatidylinositol 3-Kinase Pathway by Nobiletin, a Polymethoxy Flavonoid, Results in Augmentation of Tissue Inhibitor of Metalloproteinases-1 Production and Suppression of Production of Matrix Metalloproteinases-1 and -9 in Human Fibrosarcoma HT-1080 Cells. Cancer Res. 2002;62(4):1025–9.
Yoshimizu N, Otani Y, Saikawa Y, Kubota T, Yoshida M, Furukawa T, Kumai K, Kameyama K, Fujii M, Yano M, et al. Anti-tumour effects of nobiletin, a citrus flavonoid, on gastric cancer include: antiproliferative effects, induction of apoptosis and cell cycle deregulation. Aliment Pharmacol Ther. 2004;20:95–101.
Tominari T, Hirata M, Matsumoto C, Inada M, Miyaura C. Polymethoxy Flavonoids, Nobiletin and Tangeretin, Prevent Lipopolysaccharide-Induced Inflammatory Bone Loss in an Experimental Model for Periodontitis. J Pharmacol Sci. 2012;119(4):390–4.
Evans M, Sharma P, Guthrie N. Bioavailability of ctrus polymethoxylated flavones and their biological role in metabolic syndorome and hyperlipidemia. In: Noreddin A, editors. Readings in advanced pharmacokinetics - Theory, methods and applications. Intech; 2012. p. 267-84. http://www.intechopen.com/books/readings-in-advanced-pharmacokinetics-theory-methods-and-applications.
Li RW, Theriault AG, Au K, Douglas TD, Casaschi A, Kurowska EM, Mukherjee R. Citrus polymethoxylated flavones improve lipid and glucose homeostasis and modulate adipocytokines in fructose-induced insulin resistant hamsters. Life Sci. 2006;79(4):365–73.
Cui Y, Wu J, Jung S-C, Park D-B, Maeng Y-H, Hong JY, Kim S-J, Lee S-R, Kim S-J, Kim SJ, et al. Anti-neuroinflammatory Activity of Nobiletin on Suppression of Microglial Activation. Biol Pharm Bull. 2010;33(11):1814–21.
Fatokun AA, Liu JO, Dawson VL, Dawson TM. Identification through high-throughput screening of 4′-methoxyflavone and 3′,4′-dimethoxyflavone as novel neuroprotective inhibitors of parthanatos. Br J Pharmacol. 2013;169(6):1263–78.
Kim D, Lee K, Eun J, Zee O, Lim J, Eum S, Kim S, Shin T. Anti-allergic components from the peels ofCitrus unshiu. Arch Pharm Res. 1999;22(6):642–5.
Akao Y, Itoh T, Ohguchi K, Iinuma M, Nozawa Y. Interactive effects of polymethoxy flavones from Citrus on cell growth inhibition in human neuroblastoma SH-SY5Y cells. Bioorg Med Chem. 2008;16(6):2803–10.
Murakami A, Song M, Katsumata S-I, Uehara M, Suzuki K, Ohigashi H. Citrus nobiletin suppresses bone loss in ovariectomized ddY mice and collagen-induced arthritis in DBA/1 J mice: Possible involvement of receptor activator of NF-kappaB ligand (RANKL)-induced osteoclastogenesis regulation. Biofactors. 2007;30(3):179–92.
Joshi CP, Chiang VL. Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferase. Plant Mol Biol. 1998;37:663–74.
Meyermans H, Morreel K, Lapierre C, Pollet B, De Bruyn A, Busson R, Herdewijn P, Devreese B, Van Beeumen J, Marita JM, et al. Modifications in Lignin and Accumulation of Phenolic Glucosides in Poplar Xylem upon Down-regulation of Caffeoyl-Coenzyme A O-Methyltransferase, an Enzyme Involved in Lignin Biosynthesis. J Biol Chem. 2000;275(47):36899–909.
Zhong R, Morrison WH, Himmelsbach DS, Poole FL, Ye Z-H. Essential Role of Caffeoyl Coenzyme A O-Methyltransferase in Lignin Biosynthesis in Woody Poplar Plants. Plant Physiol. 2000;124(2):563–78.
Brunet G, Saleh Nabiel AM, Ibrahim Ragai K. Notizen: Stepwise Methylation of Quercetin by Cell-Free Extracts of Citrus Tissues. In: Zeitschrift für Naturforschung C, vol. 33. 1978. p. 786.
Brunet G, Ibrahim RK. O-methylation of flavonoids by cell-free extracts of calamondin orange. Phytochemistry. 1980;19(5):741–6.
Liu X, Luo Y, Wu H, Xi W, Yu J, Zhang Q, Zhou Z. Systematic analysis of O-methyltransferase gene family and identification of potential members involved in the formation of O-methylated flavonoids in Citrus. Gene. 2016;575(2, Part 2):458–72.
Schroder G, Wehinger E, Lukacin R, Wellmann F, Seefelder W, Schwab W, Schroder J. Flavonoid methylation: a novel 4′-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases. Phytochemistry. 2004;65(8):1085–94.
Ibrahim RK, Bruneau A, Bantignies B. Plant O-methyltransferase: molecular analysis, common signature and classification. Plant Mol Biol. 1998;36:1–10.
Kim B-G, Sung SH, Chong Y, Lim Y, Ahn J-H. Plant Flavonoid O-Methyltransferases: Substrate Specificity and Application. J Plant Biol. 2010;53(5):321–9.
Zubieta C, Kota P, Ferrer J-L, Dixon RA, Noel JP. Structural Basis for the Modulation of Lignin Monomer Methylation by Caffeic Acid/5-Hydroxyferulic Acid 3/5-O-Methyltransferase. Plant Cell. 2002;14(6):1265–77.
Zubieta C, He XZ, Dixon RA, Noel JP. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferase. Nat Struct Mol Biol. 2001;8:271–9.
Cacace S, Schröder G, Wehinger E, Strack D, Schmidt J, Schröder J. A flavonol O-methyltransferase from Catharanthus roseus performing two sequential methylations. Phytochemistry. 2003;62(2):127–37.
Zhou J-M, Gold ND, Martin VJJ, Wollenweber E, Ibrahim RK. Sequential O-methylation of tricetin by a single gene product in wheat. Biochim Biophys Acta, Gen Subj. 2006;1760(7):1115–24.
Berim A, Hyatt DC, Gang DR. A Set of Regioselective O-Methyltransferases Gives Rise to the Complex Pattern of Methoxylated Flavones in Sweet Basil. Plant Physiol. 2012;160(2):1052–69.
Willits MG, Giovanni M, Prata RT, Kramer CM, De Luca V, Steffens JC, Graser G. Bio-fermentation of modified flavonoids: an example of in vivo diversification of secondary metabolites. Phytochemistry. 2004;65(1):31–41.
Joe EJ, Kim BG, An BC, Chong Y, Ahn JH. Engineering of flavonoid O-methyltransferase for a novel regioselectivity. Mol Cells. 2010;30(2):137–41.
Wang J, Pichersky E. Identification of specific residues involved in substrate discrimination in two plant O-methyltransferases. Arch Biochem Biophys. 1999;368(1):172–80.
Dunlevy JD, Sool KL, Perkins MV, Dennis EG, Keyzers RA, Kalua CM, Boss PK. Two O-methyltransferase involved in the biosynthesis of methoxypyrazines: grape-derived aroma compounds important to wine flavour. Plant Mol Biol. 2010;74:77–89.
Frick S, Kutchan TM. Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J. 1999;17(4):329–39.
Green MR, Sambrook J. Molecular cloning : a laboratory manua. 3rd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001.
Dellaporta SL, Wood J, Hicks JB. A Plant DNA Minipreparation: Version II. Plant Mol Biol Rep. 1983;1(4):19–21.
Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25(3):674–81.
Availability of data and materials
The data supporting the results presented in this article are included as additional files. The CdFOMT gene sequences were deposited in the DDBJ (http://www.ddbj.nig.ac.jp/) under the following accession numbers: CdFOMT1, LC126056; CdFOMT3, LC126057; CdFOMT4, LC126058; CdFOMT5, LC126059; and CdFOMT6, LC126060.
Authors’ contributions
NI conceived and supervised this project and wrote the manuscript. HT conducted all experiments and wrote the manuscript. CI cooperated with HT in the characterization of the recombinant enzyme and biotransformation. All authors have read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Approval.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Primers for cloning CdFOMT-coding genes from Citrus depressa. (XLSX 14 kb)
Additional file 2: Figure S1.
Gene structure of CdFOMT sequences from Citrus depressa. Exons and introns are expressed as boxes and lines, respectively. Black boxes indicate conserved motifs that are involved in interactions with SAM in plant OMTs. Introns inserted into each gene, in which neighboring amino acid sequences are highly conserved across CdFOMTs, are marked with asterisks. Figure S2. (A) Optimum pH and (B) temperature for O-methyltransferase activity of recombinant CdFOMT5. Enzyme activity was measured in the following buffers: 50 mM sodium citrate (filled diamonds), 50 mM MES (filled squares), 50 mM phosphate-KOH (filled triangles), 50 mM Tris-HCl (filled circles), and 50 mM glycine-NaOH (multiplication sign). Enzyme activity was measured for 1 h with shaking at various temperatures. Figure S3. Mass spectrometry analysis of quercetin products methylated by CdFOMT5. The peak labeled with ‘x’ was not identified as corresponding to flavonoid derivatives. Figure S4. HPLC and LC-MS analysis of methylated products of various flavonoids by CdFOMT5. S, substrate control; S + E, enzyme reaction product; P, product peak. For equol, P1 and P2 indicate major and minor monomethylated products, respectively. Peaks labeled with ‘x’ were not identified as flavonoid derivatives. (PPTX 187 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Itoh, N., Iwata, C. & Toda, H. Molecular cloning and characterization of a flavonoid-O-methyltransferase with broad substrate specificity and regioselectivity from Citrus depressa . BMC Plant Biol 16, 180 (2016). https://doi.org/10.1186/s12870-016-0870-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-016-0870-9