Abstract
Background
Several Matrix-Assisted Laser Desorption/Ionization Time-of-Flight mass spectrometry protocols, which differ in identification criteria, have been developed for mold and dermatophyte identification. Currently, the most widely used approach is Bruker technology, although no consensus concerning the log(score) threshold has been established. Furthermore, it remains unknown how far increasing the number of spots to compare results might improve identification performance.
In this study, we used in-house and Bruker reference databases as well as a panel of 422 isolates belonging to 126 species to test various thresholds. Ten distinct identification algorithms requiring one to four spots were tested.
Results
Our findings indicate that optimal results were obtained by applying a decisional algorithm in which only the highest score of four spots was taken into account with a 1.7 log(score) threshold. Testing the entire panel enabled identification of 87.41% (in-house database) and 35.15% (Bruker database) of isolates, with a positive predictive value (PPV) of 1 at the genus level for both databases as well as 0.89 PPV (in-house database) and 0.72 PPV (Bruker database) at the species level. Applying the same rules to the isolates for which the species were represented by at least three strains in the database enabled identification of 92.1% (in-house database) and 46.6% (Bruker database) of isolates, with 1 PPV at the genus level for both databases as well as 0.95 PPV (in-house database) and 0.93 PPV (Bruker database) at the species level.
Conclusions
Depositing four spots per extract and lowering the threshold to 1.7, a threshold which is notably lower than that recommended for bacterial identification, decreased the number of unidentified specimens without altering the reliability of the accepted results. Nevertheless, regardless of the criteria used for mold and dermatophyte identification, commercial databases require optimization.
Similar content being viewed by others
Background
Until a few years ago, medical laboratories performed mold identification using phenotypic characterization of colonies and occasional sequencing of informative DNA targets. Currently, the MALDI-TOF MS (Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry) approach appears to be a promising method to identify molds and dermatophytes [1–8].
Many teams around the world have demonstrated that filamentous fungi could be identified by using MALDI-TOF MS. They first focused attention on a few specific mold and dermatophyte genera [1, 9–16]. Only after 2010 did research teams begin optimizing protocols to establish a MALDI-TOF MS-based identification approach for routine diagnosis of filamentous fungi and dermatophytes [2–7, 17–21]. Currently, four commercial systems are available for routine diagnosis, and an increasing number of publications have tested their performance in identifying molds and dermatophytes. Some teams have assessed the Saramis [12, 18, 22, 23], Vitek MS [24–28] or Andromas [29] systems; however, most publications refer to the Bruker technique [1–8, 16, 17, 19–21, 26, 30, 31]. According to the Bruker recommendations, the level of similarity between an unknown tested specimen and a reference sample is indicated by a log(score), which is referred to herein as “score”. A score > 2.3 indicates “highly probable species identification”, a score > 2 and < 2.299 indicates “secure genus identification, probable species identification”, a score > 1.7 and < 1.999 indicates “probable genus identification”, and a score < 1.7 indicates “unreliable identification”.
However, these score thresholds have been designed for bacteria identification and are not necessarily appropriate for fungi (especially molds and dermatophytes) while using the manufacturer’s reference database [4, 7, 17, 32, 33]. The majority of publications dealing with MALDI-TOF MS routine identification of filamentous fungi have used a single score threshold, usually ranging from 1.7 to 2.0, depending on the study [3, 4, 6, 8, 17]. However, a study by Shultness et al. [8] has compared the 1.7 and 2.0 score thresholds for MALDI-TOF MS identification after a liquid culture growth step in which no assessment was performed to justify these thresholds. Additionally, while the manufacturer typically recommends a single spot for bacterial identification, some authors have advocated the use of several spots of the same sample to identify molds and dermatophytes [2, 21]. According to these authors, the concordance between identification results can be tested using replicates. Theoretically, this may improve the reliability of identification results, although no study has thoroughly evaluated the optimal number of spots and log(score) threshold for mold identification. Altogether, the use of various thresholds and validation criteria for mold and dermatophyte identification complicate the interpretation of results.
The present study assessed 10 distinct identification algorithms combining several thresholds for one to four spots per sample. Because the Bruker reference database is not comprehensive enough to enable proper identification of many mold species, [4, 8] identification algorithms were assessed using an extensive in-house database (5044 references belonging to 619 species and 165 genera) and the Bruker commercial database (V3.2.1.1 : 604 references belonging to 237 species and 67 genera). Our objective was to optimize the fungal identification workflow in laboratories equipped with a Bruker MALDI-TOF MS identification platform.
Methods
Reference databases
The in-house reference database was constructed using a Microflex LT system (Bruker Daltonics, Bremen, Germany) in coordination with the BCCM/IHEM culture collection and the Mycology Laboratory of the AP-HM with the protocol described by Normand et al. [6] (several strains per species, four subcultures per strain and 10 spots per subculture, for a total of four references per strain). Details of the MS in-house database are listed in Additional file 1: Table S1.
Assessment panel
The assessment panel consisted of filamentous fungus strains that were identified via both DNA sequencing and MALDI-TOF MS. The strains were either selected at the Mycology Laboratory of Marseille (n = 224) or the BCCM/IHEM in Brussels (n = 198). Of the 224 strains from Marseille, 177 were included because the identification at the species level had to be either clarified or confirmed at the time of the study, while the remaining 47 were randomly selected among frequently identified species in the clinical laboratory. The 198 Belgian strains either originated from the BCCM/IHEM collection or were clinical strains from a previous study [34]. The 422-strain panel included 126 different species belonging to 38 genera, of which 280 strains were represented three or more times in the MALDI-TOF MS reference database (191 strains for the commercial Bruker database), 130 strains were represented once or twice (76 strains for the commercial Bruker database), and 12 strains were not represented in the reference database (155 strains for the commercial Bruker database) (Tables 1 and 2). All samples were first included in the study panel, even those corresponding to species absent from the databases to ensure that the tested algorithms correctly excluded absent species. Next, the identification process was tested again, this time only including the isolates corresponding to species represented by at least three distinct strains in the database to comply with the recommendations we have issued in a previous study [6].
Identification of the strains from the assessment panel
MALDI-TOF MS-based identification
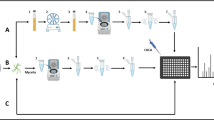
Liquid culture technology used in the Bruker protocol being not available in the two centers that participated to the study, culturing was performed on solid agar. After culturing the molds on solid Sabouraud Chloramphenicol Gentamicin agar plates (pH 5.6; glucose 40 g/L) (OXOID, Dardilly, France) for at least 48 h at 30 °C and the dermatophytes on solid Sabouraud Actidione (Bio Rad) at 27 °C, the strains were treated as described by Cassagne et al.[2], and as advised by the manufacturer : the colonies were gently scrapped using a scalpel blade, and the fungal material (approximately 2–3 mm in diameter) was suspended in a microtube containing 900 μL of anhydrous ethyl alcohol (Carlo Erba SDS, Val de Reuil, France) and 300 μL of sterile water (Water HPLC, Prolabo, BDH, Fontenay-sous-Bois, France). After a 10-min centrifugation step at 13,000 rpm, the pellet was resuspended in 10 μL of formic acid (Sigma-Aldrich, Lyon, France). After 5 min of incubation, 10 μL of acetonitrile (Prolabo BDH) was added. The suspension was then centrifuged at 13,000 rpm for 2 min, and four spots of 1 μL of protein extract per isolate were deposited. Last, the samples were covered with 1 μL of HCCA (α-cyano-4-hydroxycinnamic acid) matrix (Sigma-Aldrich, Lyon, France). MS acquisition was performed either in Marseille or Brussels using a Microflex LT system. All raw spectra were then centralized in Marseille and compared to the two reference databases. For all 1688 tested spectra (four spots for each strain) and each database, three possible scores were collected: the highest score corresponding to a true MS identification result at the species level if available, the highest score corresponding to a true identification at the genus level but false at the species level if available, and in all cases, the highest score false at the genus level.
DNA sequence-based identification
All strains belonging to the assessment panel were identified via DNA sequencing, which is the current gold standard in fungal identification. Nucleotide sequence analysis was performed either in Marseille (n = 224) or Brussels (n = 198). The rRNA ITS2 region was sequenced for each strain (using primer sequences ITS3 – GCA TCG ATG AAG AAC GCA GC and ITS4c – TCC TCC GCT TAT TGA TAT GC). An additional locus was analyzed to identify particular taxa: the partial beta-tubulin gene (primer sequences: Bt2A – GGT AAC CAA ATC GGT GCT GCT TTC and Bt2B – ACC CTC AGT GTA GTG ACC CTT GGC) was sequenced for Aspergillus, Penicillium and Scedosporium species, while the elongation factor (primer sequences: EF1 – ATG GGT AAG GAR GAC AAG AC and EF2 – GGA RGT ACC AGT SAT CAT GTT) was sequenced for Fusarium species. The hybridization temperatures applied for these primers were 54 °C for the ITS and beta-tubulin genes and 58 °C for the elongation factor gene. The DNA sequence-based identification criteria were as follows: a sequence longer than 350 base pairs and at least 99% homology with the NCBI and CBS nucleotide databases [35, 36].
Identification algorithms
The following 10 distinct algorithms were used to identify the 422 isolates included in the panel.
-
A:
Only one spot (i.e., the first of the four spots) is taken into account.
-
B:
The first two spots are taken into account.
-
B1: Only the identification corresponding to the higher of the two scores is taken into account, plotted and categorized.
-
B2: To be accepted at the species level, both identifications corresponding to the two spots must be identical, and the same rule applies for the identification at the genus level.
-
-
C:
The first three spots are taken into account.
-
C1: Only the identification corresponding to the highest of the three scores is taken into account, plotted and categorized.
-
C2: To be categorized as a concordant identification, the identification corresponding to the highest score must be identical to at least one of the other two identifications (at either the species or genus level).
-
C3: To be categorized as a concordant identification, the three identifications corresponding to the three spots must be identical (at either the species or genus level).
-
-
D:
All four spots are taken into account.
-
D1: Only the identification corresponding to the highest of the four scores is taken into account, plotted and categorized.
-
D2: To be categorized as a concordant identification, the identification corresponding to the highest score must be identical to at least one of the other three identifications (at either the species or genus level).
-
D3: To be categorized as a concordant identification, the identification corresponding to the highest score must be identical to at least two of the other three identifications (at either the species or genus level).
-
D4: To be categorized as a concordant identification, the four identifications corresponding to the four spots must be identical (at either the species or genus level).
-
Based on the individual spectrum results, 10 identification algorithms (or combinations of parameters) were tested per strain. For each spectrum, only the first identification (either true or false at the species and genus level) and the corresponding scores were collected. Each of the 10 algorithms was then tested using the identification thresholds 1.5, 1.6, 1.7, 1.8, 1.9, 2.0, 2.1, 2.2 and 2.3 for the entire panel consisting of 422 strains. The same process was applied for a reduced panel of 280 isolates that were represented at least three times in the in-house reference database and for another reduced panel of 191 isolates that were represented at least three times in the commercial database by Bruker.
Statistical analyses
An initial identification assessment was carried out by categorizing the first identification for each spot into three classes: (1) concordant at the species level with the DNA-based identification, (2) concordant at the genus level but not at the species level with the DNA-based identification, or (3) discordant at the genus level with the DNA-based identification. We then determined the best rules for accepting identification at the genus and species levels. For each threshold, algorithm, panel and database, we calculated the percentage of submitted strains that fulfilled the criteria for identification as well as the positive predictive value (PPV) of the identification result at the genus and species levels.
Results
MALDI-TOF MS-based identification
Using our in-house database to identify the 422 strains included in the panel, a clear separation was observed between the highest scores corresponding to true identifications at the species level and the highest scores corresponding to a false genus identification. Conversely, it was markedly more difficult to define a satisfying cut-off that differentiated between the highest scores corresponding to an accurate species identification and the highest scores corresponding to an accurate genus but false species identification (Fig. 1a). Using the commercial database led to a similar observation, although the separation between the three curves was less pronounced (Fig. 1b).
Highest log(score) value distribution of the 1688 tested spectra. a considering the first accurate species identification (dark line), the first accurate genus identification (gray line), and the first false genus identification (dotted line) using the in-house reference database. b considering the first accurate species identification (dark line), the first accurate genus identification (gray line), and the first false genus identification (dotted line) using the Bruker reference database
Algorithm performance and identification score threshold
Analyses of the identification performances for the 422-strain panel with the Bruker database and the in-house database. The percentage of identified strains and PPVs at the species and genus levels obtained using the 10 distinct identification algorithms were assessed on the 422-strain panel using the Bruker database (Table 3) as well as the in-house reference database (Table 4).
Notable differences in successful identification rates were highlighted between the 10 algorithms (up to 22% difference between the best identification rate and the worst one for one defined threshold). In particular, with both references databases, the algorithm that selected the best results among four spots (D1) increased the accepted identification rate (i.e., percentage of assessed strains that fulfilled the criteria for identification at either the genus or species level) (A < B1 < C1 < D1) without notably changing the identification PPV. By contrast, the inter-spot concordance criteria did not increase the identification PPV at the species level and reduced the accepted result rate (D4 < D3 < D2 ≈ D1). The main difference between the results obtained with the two databases was the percentage of accepted identifications, which was markedly lower with the Bruker database than with the in-house reference database (up to 59.8% difference between the two databases). Additionally, while the PPV for genus level identification was 100% for both databases using the 1.7 threshold, regardless of the other decision criteria applied, the PPV for species level identification was better with our in-house database than with the Bruker database (from 0.13 units for the D4 algorithm to 0.18 units for the A1 and B1 algorithms). Using the in-house reference database, the D1 combination with a threshold at 1.7 yielded 87.41% of accepted identifications, with a species identification PPV of 0.89.
We noted an increase in the species PPV with threshold values higher than 1.7, but the loss in percentage of identification is important. Using the identification score threshold of 2.00, as recommended by the manufacturer, led to a 23.75% decrease in accepted identifications and only a 2% increase in the PPV. The “highly probable species identification” criterion by Bruker (i.e., a score above the threshold of 2.3) yielded a species PPV of 0.92 but only 18.05% of accepted identifications (76 strains) using our in-house database. The identification results were even less acceptable when using the commercial database, with only 44 strains (10.45%) reaching the 2.0 threshold and 5 strains (1.19%) reaching the 2.3 threshold.
Description of the misidentifications obtained with both databases
For 32 strains, the identification best score using the D1 algorithm was higher than the 1.7 threshold, although the species level result was incorrect using the in-house database. For all of these strains, proper identification at the species level was impeded by the close similarity of the MALDI-TOF MS spectra of some species belonging to the same species complex (as it is the case with Fusarium solani and its closely related species Fusarium petrolophilum). The details of these strains are presented in Table 5. The same finding was observed for 39 misidentifications obtained using the Bruker database (Table 6).
Analyses of the identification performances for the restricted panels with the Bruker database and the in-house database
The robustness of the identification algorithms was tested by performing the same analysis on a restricted panel including only the strains that were represented by at least three distinct strains in the reference spectrum libraries (the restricted panel used with the Bruker database (n = 191) is different than the one used with the in-house database (n = 280) due to the differences in databases composition). As expected, the results were better than those obtained with the 422-strain panel (Tables 7 and 8), with a higher percentage of results reaching the thresholds (from 3.30% increase in percentage of accepted identification with B1 to 24.94% increase with D4 when selecting the 1.7 threshold) and a reduced number of misidentifications. For example, using the in-house database, the species PPV for the D1 algorithm with a 1.7 score threshold on the comprehensive 422-strain panel increased from 0.89 to 0.97 when testing a sub-panel including only the 280 strains for which species were represented by at least three distinct strains. Similar trends were observed when using the commercial database (increase in the species PPV from 0.72 to 0.93).
Twenty-two species (180 isolates) of this study are represented by at least three references in both databases. We performed a per species comparison of the efficiency of the two databases for those 22 species, using the D1 algorithm. Results are compiled in Table 9. For the total of the 180 isolates, the percentage of accepted identifications is lower with the Bruker database than with the in-house database (47.22% vs 91.67%). The comparison of the species PPV obtained with both databases highlighted weaknesses for four species (A. alternata, F. solani, P. chrysogenum and S. apiospermum) with the in-house database (due to misidentifications with closely related species), and for five species (A. parasiticus, F. proliferatum, S. brevicaulis, T. interdigitale and T. rubrum) with the Bruker database. The species PPV was equal to 1.00 and identical between the two databases for 13 species of this group.
Discussion
As shown in this study and several others, MALDI-TOF MS has recently been optimized to identify filamentous fungi at the species level, provided that an appropriate database is available [2, 4, 6–8]. The assessment panel used in this study comprised isolates belonging to 126 different species, including many difficult-to-identify isolates. Consequently, the identification rate was low compared with previously published studies, especially studies utilizing the commercial database. Using such a challenging panel, it was possible to define a threshold discriminating correct identifications at the species level from false identifications at the genus level, although identifying a threshold that differentiated between a correct and false identification at the species level among closely related species was not feasible. Our findings indicate that the best identification results (PPV at the species level and percentage of accepted identifications) were obtained when applying a decisional algorithm in which only the highest score of four spots was taken into account, and the identification was accepted if the score was at least 1.7. This threshold is notably lower than that recommended for bacteria identification, and has already been advised by several authors for yeast identification [32, 33]. Furthermore, we also observed a marked decrease in percentage of identification between a threshold of 1.7 and 2.0. When the threshold was set too high, we noted a 20% loss in identification efficiency and only a small benefit of 2% in the differentiation of certain close species. Even if the Bruker commercial database yielded a markedly lower percentage of accepted identifications (probably due to the fact that the database was built using liquid culture media, that is does not frequently involve more than two or three references per species, and that there are errors of reference labeling), the PPV remained correct for the spectra reaching the 1.7 threshold. This result corroborates the findings of several authors who have suggested that lowering the identification score thresholds would improve MALDI-TOF MS-based fungal identification efficiency [7, 17, 32, 33]. However, we strongly advise against a threshold below 1.7, which could yield an increase in false identifications, in particular at the genus level, as shown in Tables 3 and 4.
The major problem encountered with the data set obtained from the assessment panel was the resemblance between the MALDI-TOF MS spectra of some species. For 32 strains, the similarity in MALDI-TOF MS spectra yielded an incorrect identification at the species level, although these identification errors systematically corresponded to taxonomically close species often belonging to the same species complex (except for Pleospora papaveracea, which was identified as Ulocladium oudemansi). Among these 32 strains (27 species), only a few species are known to be pathogenic to humans, of which none are major human pathogens. Regarding the pathogenic species that were misidentified in our study, the treatment would not vary from that prescribed based on molecular identification, and the differentiation of these species is not clinically relevant. The remaining species are often considered to be a result of contamination, and their antifungal susceptibility is unknown. Interestingly, raising the identification score threshold to 2.3 did not entirely resolve this identification issue of taxonomically close species, as six strains were incorrectly characterized despite a score above 2.3, even using our extensive in-house database. Thus, in our study, one could question the relevance of capacity and necessity to distinguish between these closely related species as none of the misidentifications implied a change in treatment.
Our results also show that the key step in efficient identification was to increase the number of replicate spots per strain. Testing four spots of each filamentous fungus yielded better identification performance, with an increase of 5% to 7% of accepted strain identifications. Such spot replications did not markedly increase the workload, as it took only a few seconds to perform the additional spots. However, the replicates may increase the per-strain acquisition time in the laboratory workflow. The four-spot method has been applied in the routine workflow at the Marseille Mycology Laboratory for the past five years, which demonstrates that this procedure is easily applicable in hospital laboratories to identify filamentous fungi isolated from clinical samples and from hospital environment samples. Multiplying the number of deposits per strains imply a small increase in the cost per strain as only one reagent (HCCA matrix) has to be used more extensively. The decisional algorithm implying concordance of the identification results of two to four spots did not enhance identification performance, regardless of the threshold selected. Furthermore, the D2, D3 and D4 algorithms tended to lower the percentage of validated identifications by disregarding identification results that would have been correct using the D1 decisional algorithm.
When focusing on the species that were represented by at least three distinct strains in the reference spectrum library, identification efficacy markedly increased. This result stresses the importance of a comprehensive reference spectrum library for the identification of filamentous fungi, including dermatophytes, as previously reported by our group [6], and as previously demonstrated for yeasts [32]. The multiplication of strains per species as well as subcultures per strain allowed us to cover the variations seen within the environment for filamentous fungi.
Our in-house reference database, which yielded markedly better results compared with the commercial Bruker database, was constructed mainly with collection strains and with some clinical strains isolated in our laboratory and obtained during routine analyses. These clinical strains were later included in the BCCM/IHEM collection in Brussels. Nevertheless, even our enhanced database still requires optimization to identify rare species. Indeed, the poor representation of rare species in the database may explain the identification percentage of 87.4% obtained with this challenging panel of 422 strains (92.1% with the 280 strains represented by more than three strains in the reference database), compared with the extremely good identification results of common species (98.1%) exposed by Gautier et al. [21].
Conclusion
The growing number of publications on MALDI-TOF MS-based fungal identification indicates that this technology is widely used and applicable to the routine laboratory workflow. MALDI-TOF MS-based identification of fungi yields exceedingly more accurate results compared with morphology-based analyses [21]. Furthermore, this technology is less expensive and easier than the current DNA based-identification gold standard, with a turnaround time allowing achieving the analysis more rapidly. Currently, the major limitation of this approach is the lack of a comprehensive and efficient reference spectrum library. We acknowledge that in-house reference databases that are non-open-source, such as ours, are of little benefit to the global scientific community. Therefore, we plan to deploy an accessible version of our database along with an identification algorithm that can be queried online to assist scientific teams to expand the identification of fungal species. Experiments implying a multicenter approach are ongoing to finalize this project.
References
Marinach-Patrice C, Lethuillier A, Marly A, Brossas J-Y, Gené J, Symoens F, Datry A, Guarro J, Mazier D, Hennequin C. Use of mass spectrometry to identify clinical Fusarium isolates. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2009;15:634–42.
Cassagne C, Ranque S, Normand A-C, Fourquet P, Thiebault S, Planard C, Hendrickx M, Piarroux R. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One. 2011;6:e28425.
De Carolis E, Posteraro B, Lass-Flörl C, Vella A, Florio AR, Torelli R, Girmenia C, Colozza C, Tortorano AM, Sanguinetti M, Fadda G. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2012;18:475–84.
Lau AF, Drake SK, Calhoun LB, Henderson CM, Zelazny AM. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:828–34.
L’Ollivier C, Cassagne C, Normand A-C, Bouchara J-P, Contet-Audonneau N, Hendrickx M, Fourquet P, Coulibaly O, Piarroux R, Ranque S. A MALDI-TOF MS procedure for clinical dermatophyte species identification in the routine laboratory. Med Mycol. 2013;51:713–20.
Normand A-C, Cassagne C, Ranque S, L’ollivier C, Fourquet P, Roesems S, Hendrickx M, Piarroux R. Assessment of various parameters to improve MALDI-TOF MS reference spectra libraries constructed for the routine identification of filamentous fungi. BMC Microbiol. 2013;13:76.
Ranque S, Normand A-C, Cassagne C, Murat J-B, Bourgeois N, Dalle F, Gari-Toussaint M, Fourquet P, Hendrickx M, Piarroux R. MALDI-TOF mass spectrometry identification of filamentous fungi in the clinical laboratory. Mycoses. 2014;57:135–40.
Schulthess B, Ledermann R, Mouttet F, Zbinden A, Bloemberg GV, Böttger EC, Hombach M. Use of the Bruker MALDI Biotyper for identification of molds in the clinical mycology laboratory. J Clin Microbiol. 2014;52:2797–803.
Li TY, Liu BH, Chen YC. Characterization of Aspergillus spores by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom RCM. 2000;14:2393–400.
Welham KJ, Domin MA, Johnson K, Jones L, Ashton DS. Characterization of fungal spores by laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom RCM. 2000;14:307–10.
Chen H-Y, Chen Y-C. Characterization of intact Penicillium spores by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom RCM. 2005;19:3564–8.
Dong H, Kemptner J, Marchetti-Deschmann M, Kubicek CP, Allmaier G. Development of a MALDI two-layer volume sample preparation technique for analysis of colored conidia spores of Fusarium by MALDI linear TOF mass spectrometry. Anal Bioanal Chem. 2009;395:1373–83.
Kemptner J, Marchetti-Deschmann M, Mach R, Druzhinina IS, Kubicek CP, Allmaier G. Evaluation of matrix-assisted laser desorption/ionization (MALDI) preparation techniques for surface characterization of intact Fusarium spores by MALDI linear time-of-flight mass spectrometry. Rapid Commun Mass Spectrom RCM. 2009;23:877–84.
Tao J, Zhang G, Jiang Z, Cheng Y, Feng J, Chen Z. Detection of pathogenic Verticillium spp. using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom RCM. 2009;23:3647–54.
DE Respinis S, Vogel G, Benagli C, Tonolla M, Petrini O, Samuels G. MALDI-TOF MS of Trichoderma: A model system for the identification of microfungi. Mycol Prog. 2010;9:79–100.
Coulibaly O, Marinach-Patrice C, Cassagne C, Piarroux R, Mazier D, Ranque S. Pseudallescheria/Scedosporium complex species identification by Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry. Med Mycol Off Publ Int Soc Hum Anim Mycol. 2011;49:621–6.
Del Chierico F, Masotti A, Onori M, Fiscarelli E, Mancinelli L, Ricciotti G, Alghisi F, Dimiziani L, Manetti C, Urbani A, Muraca M, Putignani L. MALDI-TOF MS proteomic phenotyping of filamentous and other fungi from clinical origin. J Proteomics. 2012;75(11):3314–30.
Nenoff P, Erhard M, Simon JC, Muylowa GK, Herrmann J, Rataj W, Gräser Y. MALDI-TOF mass spectrometry - a rapid method for the identification of dermatophyte species. Med Mycol. 2013;51:17–24.
Packeu A, Hendrickx M, Beguin H, Martiny D, Vandenberg O, Detandt M. Identification of the Trichophyton mentagrophytes complex species using MALDI-TOF mass spectrometry. Med Mycol. 2013;51:580–5.
Packeu A, De Bel A, l’Ollivier C, Ranque S, Detandt M, Hendrickx M. Fast and accurate identification of dermatophytes by matrix-assisted laser desorption ionization-time of flight mass spectrometry: validation in the clinical laboratory. J Clin Microbiol. 2014;52:3440–3.
Gautier M, Ranque S, Normand A-C, Becker P, Packeu A, Cassagne C, L’Ollivier C, Hendrickx M, Piarroux R. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: revolutionizing clinical laboratory diagnosis of mould infections. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2014;20:1366–71.
Erhard M, Hipler U-C, Burmester A, Brakhage AA, Wöstemeyer J. Identification of dermatophyte species causing onychomycosis and tinea pedis by MALDI-TOF mass spectrometry. Exp Dermatol. 2008;17:356–61.
de Respinis S, Tonolla M, Pranghofer S, Petrini L, Petrini O, Bosshard PP. Identification of dermatophytes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Med Mycol. 2013;51:514–21.
Verwer PEB, van Leeuwen WB, Girard V, Monnin V, van Belkum A, Staab JF, Verbrugh HA, Bakker-Woudenberg Ia JM, van de Sande WWJ. Discrimination of Aspergillus lentulus from Aspergillus fumigatus by Raman spectroscopy and MALDI-TOF MS. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2014;33:245–51.
De Respinis S, Monnin V, Girard V, Welker M, Arsac M, Cellière B, Durand G, Bosshard PP, Farina C, Passera M, Van Belkum A, Petrini O, Tonolla M. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry using the Vitek MS system for rapid and accurate identification of dermatophytes on solid cultures. J Clin Microbiol. 2014;52:4286–92.
Lévesque S, Dufresne PJ, Soualhine H, Domingo M-C, Bekal S, Lefebvre B, Tremblay C. A Side by Side Comparison of Bruker Biotyper and VITEK MS: Utility of MALDI-TOF MS Technology for Microorganism Identification in a Public Health Reference Laboratory. PLoS One. 2015;10:e0144878.
Iriart X, Lavergne R-A, Fillaux J, Valentin A, Magnaval J-F, Berry A, Cassaing S. Routine identification of medical fungi by the new Vitek MS matrix-assisted laser desorption ionization-time of flight system with a new time-effective strategy. J Clin Microbiol. 2012;50:2107–10.
McMullen AR, Wallace MA, Pincus DH, Wilkey K, Burnham CA. Evaluation of the Vitek MS Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry System for Identification of Clinically Relevant Filamentous Fungi. J Clin Microbiol. 2016;54:2068–73.
Alshawa K, Beretti J-L, Lacroix C, Feuilhade M, Dauphin B, Quesne G, Hassouni N, Nassif X, Bougnoux M-E. Successful identification of clinical dermatophyte and Neoscytalidium species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50:2277–81.
Alanio A, Beretti J-L, Dauphin B, Mellado E, Quesne G, Lacroix C, Amara A, Berche P, Nassif X, Bougnoux M-E. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for fast and accurate identification of clinically relevant Aspergillus species. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2011;17:750–5.
Theel ES, Hall L, Mandrekar J, Wengenack NL. Dermatophyte identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49:4067–71.
Vlek A, Kolecka A, Khayhan K, Theelen B, Groenewald M, Boel E, Multicenter Study Group, Boekhout T. Interlaboratory comparison of sample preparation methods, database expansions, and cutoff values for identification of yeasts by matrix-assisted laser desorption ionization-time of flight mass spectrometry using a yeast test panel. J Clin Microbiol. 2014;52:3023–9.
Van Herendael BH, Bruynseels P, Bensaid M, Boekhout T, De Baere T, Surmont I, Mertens AH. Validation of a modified algorithm for the identification of yeast isolates using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS). Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2012;31:841–8.
Becker PT, de Bel A, Martiny D, Ranque S, Piarroux R, Cassagne C, Detandt M, Hendrickx M. Identification of filamentous fungi isolates by MALDI-TOF mass spectrometry: clinical evaluation of an extended reference spectra library. Med Mycol. 2014;52:826–34.
Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, Cardinali G, Arthur I, Normand A-C, Giraldo A, da Cunha KC, Sandoval-Denis M, Hendrickx M, Nishikaku AS, de Azevedo Melo AS, Merseguel KB, Khan A, Parente Rocha JA, Sampaio P, da Silva Briones MR, e Ferreira RC, de Medeiros Muniz M, Castañón-Olivares LR, Estrada-Barcenas D, Cassagne C, Mary C, Duan SY, Kong F, Sun AY, Zeng X, et al. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database—the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med Mycol. 2015;53:313–37.
Balajee SA, Borman AM, Brandt ME, Cano J, Cuenca-Estrella M, Dannaoui E, Guarro J, Haase G, Kibbler CC, Meyer W, O’Donnell K, Petti CA, Rodriguez-Tudela JL, Sutton D, Velegraki A, Wickes BL. Sequence-Based Identification of Aspergillus, Fusarium, and Mucorales Species in the Clinical Mycology Laboratory: Where Are We and Where Should We Go from Here? J Clin Microbiol. 2009;47:877–84.
Acknowledgements
We especially thank Sandy Moore for proofreading the English version of the manuscript.
Funding
None.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
ACN and PB collected the data. ACN performed the analyses. ACN and RP interpreted the data. ACN, RP, PB, SR and MH were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Table S1.
The species included in the in-house reference database (as labeled by the BCCM/IHEM or Marseille Mycology Laboratory staff at the time of database development). Nb str: number of strains per genus; nb sp: number of species belonging to each genus. The number in brackets represents the number of strains corresponding to each species. (DOCX 52 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Normand, AC., Cassagne, C., Gautier, M. et al. Decision criteria for MALDI-TOF MS-based identification of filamentous fungi using commercial and in-house reference databases. BMC Microbiol 17, 25 (2017). https://doi.org/10.1186/s12866-017-0937-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-017-0937-2