Abstract
Background
Chloroplast plays a vital role in plant development and growth. The pentatricopeptide repeat (PPR) gene family is one of the largest gene families in plants. In addition, cold stress affects a broad spectrum of cellular components, e.g. chloroplast, and metabolism in plants. However, the regulatory mechanism for rice PPR genes on chloroplast development still remains elusive under cold stress.
Result
In this paper, we characterized a new rice PPR gene mutant tcd10 (thermo-sensitive chlorophyll-deficient mutant 10) that exhibits the albino phenotype, malformed chloroplast and could not survive after the 5-leaf stage when grown at 20 °C, but does the normal phenotype at 32 °C. Map-based cloning, followed by RNA interference and CRISPR/Cas9 genome editing techniques, revealed that TCD10 encoding a novel PPR protein, mainly localized to the chloroplasts, with 27 PPR motifs, is responsible for the mutant phenotype. In addition, TCD10 is specific expression in tissues. The disruption of TCD10 resulted in an evidently reduced expression of chloroplast-associated genes under cold stress (20 °C), whereas they did recovered to normal levels at high temperature (32 °C). These results showed an important role of TCD10 for chloroplast development under cold stress.
Conclusions
The TCD10 encodes a novel rice PPR protein, mainly located in chloroplasts, which is important for chloroplast development, growth and the maintenance of photosynthetic electron transport and its disorder would lead to an aberrant chloroplast and abnormal expressions in these genes for chloroplast development and photosynthesis in rice under cold stress.
Similar content being viewed by others
Background
The formation of normal chloroplasts is crucial for photosynthesis and carbon assimilation in plants. Chloroplast is a semi-autonomous organelle which has its own DNA genome and protein synthesis system, but the only part of the proteins can be synthesized in the chloroplast, the vast majority of proteins are synthesized on the ribosomes of the cytoplasm. Chloroplasts are mostly allocated in the leaves of plants, and its development is closely linked with the leaf development. As known, plastid development from proplastids to mature chloroplasts can be divided into three phases (Chory et al. 1991; Mullet 1993; Kusumi et al. 1997 and 2014). The first phase concerns the activation of plastid replication and plastid DNA synthesis. The second phase is the chloroplast “build-up” stage, which is characterized by the construct of the chloroplast genetic system. At this phase, the nuclear-encoded plastid RNA polymerase (NEP) preferentially transcribes the plastid genes and transcription/translation in the chloroplasts increase strikingly (Hajdukiewicz et al. 1997). In the final phase, the plastid- and nuclear genes for photosynthetic apparatus are massively expressed and principally transcribed by the plastid-encoded RNA polymerase (PEP) (De Santis-Maciossek et al. 1999). Plastid-encoded proteins assemble with imported nuclear-encoded proteins to form photosynthetic and metabolic complexes in chloroplasts. Thus, chloroplast development needs tightly coordinated gene expression between the chloroplast- and nuclear genomes.
Pentatricopeptide repeat (PPR) proteins are one of the largest protein families in plants. The PPR family is characterized by tandem arrays of the PPR motif, a highly degenerate repeat of 35 canonical amino acids (Small and Peeters 2000). PPR proteins harbor between two and 30 PPR motifs, and their tandem alignment allows the modular recognition of specific RNA sequences, generally function in chloroplast or mitochondria (Fujii et al. 2013). Recent studies showed that PPR proteins can promote versatile processes involving RNA, such as editing, splicing, stabilization, and translation (Schmitz-Linneweber and Small 2008; Fujii and Small 2011; Nakamura et al. 2012). In chloroplasts, some PPR proteins have been identified to participate in RNA splicing (Schmitz-Linneweber et al. 2006; De Longevialle et al. 2007; Ichinose et al. 2012), RNA processing (Meierhoff et al. 2003; Hattori et al. 2007), RNA editing (Kotera et al. 2005; Okuda et al. 2007; Chateigner-Boutin et al. 2008; Cai et al. 2009; Yu et al. 2009; Zhou et al. 2009; Tseng et al. 2010; Sosso et al. 2012), translation (Williams and Barkan 2003; Tavares-Carreón et al. 2008), and RNA stability (Yamazaki et al. 2004; Pfalz et al. 2009; Fujii et al. 2013). Despite the fact, a lot of work still to be done is to discriminate the functions of PPR genes in plant development, especially in rice.
Rice (Oryza sativa) is the most important food crop and has also been established as a model species for plant genome research. It is known that a mutation in a PPR gene usually has a strong phenotypic effect (Kocábek et al. 2005). Notably, functional studies of rice PPR proteins remain much sparse. OsPPR1, carrying 11 PPR motifs, is the first identified PPR protein responsible for rice chloroplast development (Gothandam et al. 2005). Antisense transgenic approach was exploited to suppress the expression of OsPPR1 and the obtained transgenic lines displayed chlorophyll-deficiency, albinism and lethality. Another rice PPR protein, YSA, with 16 PPR motifs, also is essential for chloroplast development and its disruption causes a seedling stage-specific albino phenotype (Su et al. 2012). Also the chloroplast-localized PPR protein OsV4 is needed for chloroplast development at early seedling stage under cold stress (Gong et al. 2014). More recently, also we reported that rice ASL3 with 10 PPR motifs is required for chloroplast development and its disruption lead to the death of seedlings (Lin et al. 2015a, b). Herein, we describe a novel thermo-sensitive chlorophyll-deficient mutant, tcd10, which displayed the albino phenotype below 20 °C and TCD10 encodes a novel PPR protein, containing 27 PPR motifs, required for chloroplast development and photosynthesis in rice under cold stress.
Results
Phenotypic Analysis of the tcd10 Mutant
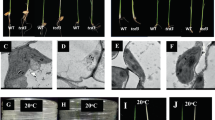
The color of the tcd10 mutant seedlings grown at 20 °C, 24 °C, 28 °C and 32 °C, respectively, was shown in Fig. 1a. Apparently, the mutant seedlings displayed albino phenotype, thereafter could not survive past 5-leaf stage at 20 °C(data not shown); the degree of chlorosis gradually weakened when grown at 24 °C and 28 °C, but become green as WT plants at 32 °C, indicating the low thermo-sensitivity of the mutant phenotype. Consist with the observed phenotype, the accumulations of chlorophyll a, b, and carotenoid in tcd10 plants at 20 °C (Fig. 1b) were drastically lower than those at 32 °C and WT plants (Fig. 1c). It is indicated that the low temperature led to the block of formation of photosynthetic pigments in tcd10 mutants.
Characterization of the tcd10 mutants; a 4-leaf stage seedlings of wild type (WT) (right) and tcd10 mutants (left) grown at 20 °C, 24 °C, 28 °C and 32 °C, respectively; b, c photosynthetic pigment contents of tcd10 and WT 3-leaf stage seedlings at 20 °C and 32 °C respectively; Chl, total chlorophyll; Chla, chlorophyll a; Chlb, chlorophyll b; Car, carotenoid
The alterations of leaf-color are generally accompanied with the development of chloroplast. To verify this view, we compared the chloroplast ultrastructure of tcd10 and WT leaves under 32 °C and 20 °C. As expected, WT cells (Fig. 2) under 32 °C and 20 °C contained normal chloroplasts with well-organized lamellar structures and were equipped with normally stacked grana and thylakoid membranes. By contrast, most cells under 20 °C in tcd10 mutants contained fewer chloroplasts (Fig. 2g), which were theteroplastidic and contained non-pigmented plastids with severely vacuolated and lacked organized lamellar (Fig. 2h). However, the mutant cells at 32 °C had not obvious difference with WT plants (Fig. 2c, d). Accordingly, the tcd10 aberrant chloroplast induced by low temperatures caused the decreased accumulation of photosynthetic pigments, so as to produce the mutant phenotype.
We investigated the Fv/Fm values, reflecting the maximum potential capacity of the PSII photochemical reactions in chloroplasts (Krause and Weis 1991). Resultantly, at 20 °C, the Fv/Fm value was 0.834 ± 0.04 in WT plants, and undetectable in tcd10 mutants, indicating that the photochemical efficiency of PSII was completely hindered in tcd10 mutants under cold stress. In addition, the max rate (molm−2s−1) of photosynthetic electron transport (rETR max ) was 63.34 ± 0.484 in WT plants and undetectable in tcd10 mutants, showing that the tcd10 mutation completely blocked the electron transport under cold stress. By contrast, at 32 °C, all values in the tcd10 mutants (0.559 ± 0.012 for Fv/Fm and 157.200 ± 1.131 for rETR max , respectively) were nearly similar to WT levels (0.642 ± 0.024 for Fv/Fm and 143.800 ± 8.707 for rETR max , respectively), showing that both PSII activity and photosynthetic electron transport in the tcd10 mutants were nearly recovered to WT levels at 32 °C.
In addition, the leaf chlorophyll SPAD values (Additional file 1: Figure S1) in tcd10 plants had not significant difference from WT plants after transplanting under nature conditions. However, in contrast to have not great influence on both the final plant height (Additional file 2: Figure S2) and 1000-grain weight (Additional file 3: Figure S3), the panicle number and panicle length was significantly reduced (Additional file 3: Figure S3). These results illustrated the tcd10 mutation affects not only chloroplast development under cold stress and but also the late growth of rice under nature conditions.
Map-based Cloning of TCD10
The genetic analysis (Additional file 4: Table S1) showed that this mutation in tcd10 is a single recessive locus based on the approximate 3(green):1(albino) ratio for phenotypic segregation using F2 population generated from Guangzhan63S/tcd10. To this end, we randomly selected the 222 F2 individuals with mutant phenotype and mapped TCD10 between RM475 and RM33 on chromosome 10 (Fig. 3a). To further fine-map TCD10, a total of 5200 F2 mutant invididuals and developed five InDel molecular markers (Additional file 5: Table S2) were exploited. Ultimately, the TCD10 was localized to 70 kb between ID14874 and ID14944 (Fig. 3b). Using Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/#search), the target genomic region contained six predicted candidate genes (Fig. 3c). By sequencing and analyzing all the candidate genes, compared with wild type, in the tcd10 mutants we only uncovered a 1-bp deletion (A) at position 766 bp from the ATG start codon in the extron of LOC_Os10g28600 (Fig. 4d) which caused pre-termination, suggesting that TCD10 is LOC_Os10g28600.
Genetic analysis and cloning of the TCD10 gene; a TCD10 was located between the RM25441 and RM333 on chromosome 10 using 222 F2 mutant individuals; b TCD10 was narrowed to a 70 kb between ID14874 and ID14944 on AC027662 BAC clone using 5200 F2 mutant individuals; c The target region contains six predicted candidate genes (LOC_Os10g28590, LOC_Os10g28600 LOC_Os10g28610, LOC_Os10g28620, LOC_Os10g28630, LOC_Os10g28640); d A single nucleotide (A) deletion mutation at position 766 bp from the ATG start codon in LOC_Os10g28600; e Chloroplast transit peptide (CTP), PPR motif and mutation sites; f Phenotypes of CRISPR/Cas9 genome editing (T1-1 and T1-2) and RNAi (T1-3) transgenic lines grown at 20 °C; g Transcript levels of three RNAi (T1-3) transgenic lines
Phylogenic analysis of TCD10; a Amino acid sequence alignment of TCD10 with the five homologs. Amino acids fully or partially conserved are shaded black and gray, respectively. The homologous comparison is based on a multiple sequence with alignment algorithm MAFFT and generated with the programDNAMAN8; b Phylogenic tree of TCD10 and homologs. Protein sequences are Ricinus communis (RCOM_ 8274176), Populus trichocarpa (POPTR_0006s28060), Arabidopsis thaliana (AT4G31850), Glycine max (100778358), Vitis vinifera (100247529), Brachypodium distachyon (100836501), Sorghum bicolor (SORBI_01g032160), Zea mays (GRMZM2G372632), Selaginella moellendorffii (SELMODRAFT_76934). Scale represents percentage substitution per site. The rooted tree is based on a multiple sequence with alignment algorithm MAFFT and generated with the program Mega6
Knock-down and -out of TCD10 Exhibits the Mutant Phenotype
To further affirm that the malfunction of TCD10 is blame for the mutant phenotype, we used both RNA interference (RNAi) and CRISPR/Cas9 genome editing system to knockdown and knockout LOC_Os10g28600 in WT plants, respectively. In RNAi study, we obtained eleven-one RNAi lines all showing the chlorosis phenotype similar to tcd10 at 20 °C(Fig. 3f). Thus, the down-regulated TCD10 expression in RNAi lines (Fig. 3g) could mimic the phenotype of the tcd10 mutant. In CRISPR/Cas9 study, we obtained 38 transgenic T0 plants carrying two different editing sites (the 1-bp (G) and 17-bp (CCATGGCCGGGTCGGGG) deletion happened at position 316 bp and 306 bp from the ATG start codon in TCD10, respectively) (Additional file 6: Figure S4). Importantly, all transgenic T1 plants appeared the separation of albino phenotype as tcd10 mutants at 20 °C (Fig. 3f). Taken together, these results reconfirmed that LOC_Os10g28600 is TCD10.
Characterization of TCD10 Protein
Determination of the nucleotide sequence of the RT-PCR product revealed that TCD10 encodes a 3790 bp open-reading frame, which is not interrupted by any intron and encodes a 1080-amino acid polypeptide with a calculated molecular mass of 120kD. The TCD10 contains 27 PPR motifs of the protein (Fig. 3e, Additional file 7: Figure S5). In addition, the first 50 amino acids (Additional file 7: Figure S5) were predicted to be a chloroplast transit peptide (CTP) by targetp (http://www.cbs.dtu.dk/services/TargetP/). In addition, the two editing deletion mutations in the transgenic lines (T1-1and T1-2) and the tcd110 mutation occurred in the 1st and 5th PPR motif, respectively, and all caused the premature termination of translation, finally leading to the complete damage of protein structure (Fig. 3e, Additional file 7: Figure S5). Multiple amino acid sequence alignment showed that TCD10 was similar to many species (Fig. 4a), including Arabidopsis At4g31850/PGR3 with 62.3% of identity (Shikanai et al. 1999; Yamazaki et al. 2004; Cai et al. 2011; Fujii et al. 2013). The subsequent phylogenetic analysis showed that TCD10 had the closest genetic relationship with one of the predicted proteins in sorghum (Fig. 4b), indicating the high conservation of TCD10 protein in plants.
TCD10 Expression Pattern
To determine expression pattern of TCD10 gene in various tissues, we analyzed the TCD10 expression level of the root, stems, young-leaf, flag-leaf and panicle in wild-type plant tissues. As shown in Fig. 5a, TCD10 in all various tissues were expressed, but in the leaves was relatively higher than that of other tissues, especially in the flag leaf expression was the highest, and expression in roots and stems rarely, which basically is consistent with RiceXPro database prediction results (http://ricexpro.dna.affrc.go.jp/) (Additional file 8: Figure S6). In addition, the expression levels were increased together with leaf age (Fig. 5b), indicating the importance of TCD10 for leaf chloroplast development under cold stress.
Expression patterns of TCD10 by RT-PCR analysis; a Expression of various tissues; YR, young-seedling roots; YS, young-seedling stem; YL, young-seedling leaf; SL, second leaf; FL, flag leaf at heading; PN, young panicles. OsActin was used as a control (cycle number for OsActin was 28, cycle number for TCD10 was 35); b Expression levels at different leaves
The Transcript Expressions of the Associated Genes in the tcd10 Mutants
We investigated the transcript levels of twenty-five genes (Additional file 9: Table S3) associated with Chl biosynthesis, photosynthesis and chloroplast development (CAO1, PORA, YGL1, Cab1R, Cab2R, rbcL, rbcS, psaA, psbA, LhcpII, rps7, OsV4, rpoC, OsRpoTp, V1, V2, RNRL, RNRS, 16SrRNA, rps20, FtsZ, rpl21, and OsDG2) in the tcd10 mutants. As a result, under cold stress (20 °C) (Fig. 6a), except for the slight increase in YGL1 encoding Chl synthase, compared with WT plants, the transcripts in those genes for Chl biosynthesis, such as chlorophyllide a oxygenase (CAO1), and NADPH: protochlorophyllide oxidoreductase (PORA), were significantly impeded in tcd10 mutants, consistent with the reduced Chl content (Fig. 1c) and the mutant phenotype (Fig. 1a). Also all the seven photosynthesis-associated genes studied (Cab1R, Cab2R, rbcL, rbcS, psaA, psbA, LhcpII) (Fig. 6b) were severely hampered in tcd10 mutants, consistent with the undetectable photochemical efficiency of PSII and photosynthetic electron transport in photosystems in tcd10 mutants, thereafter led to the seedling death. Moreover, all transcript accumulations of sixteen genes tested for chloroplast development were abnormally influenced in tcd10 mutants, especially OsV4, OsRpoTp, V1, V2, RNRL, RNRS, 16SrRNA, rpl21 and OsDG2 were considerably down-regulated (Fig. 6c). More notably, at 32 °C, the transcript levels of the down-regulated genes at low temperatures in tcd10 mutants were recovered to or even slightly higher than WT plants (Fig. 7a, b, c). Accordingly, under high temperatures (32 °C), the tcd10 mutation has very little effect on the associated-genes for Chl biosynthesis, photosynthesis and chloroplast development, consistent with the normal phenotype in tcd10 mutants at 32 °C. Taken together, TCD10 is needed for chloroplast development under cold stress.
Transcript levels of those genes associated with Chl biosynthesis (a), photosynthesis (b) and chloroplast development (c) at 20 °C in WT and tcd10 mutants in the 3rd leaves, respectively. The relative expression level of each gene in WT and mutant was analyzed by qPCR and normalized using the OsActin as an internal control. Data are means ± SD (n = 3)
qPCR analysis of those genes associated with Chl biosynthesis, photosynthesis and chloroplast development at 32 °C in the 3rd leaves; a, b, c Expression levels of genes related to Chl biosynthesis (a), photosynthesis (b) and chloroplast development (c) in WT and tcd10 mutants in the 3rd leaves, respectively
Subcellular Localization of TCD10
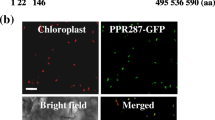
The majority of PPR proteins are predicted to be targeted to either mitochondria or chloroplasts (Small and Peeters 2000). In this study, TargetP (Emanuelsson et al. 2000, http://www.cbs.dtu.dk/services/TargetP/) and iPSORT (http://ipsort.hgc.jp/) all suggested possible chloroplast localization and secretory channels. To assess possible localization of TCD10, we constructed the recombinant expression vectors pMON530-GFP-TCD10 and transformed into tobacco cells by Agrobacterium-mediated infection. Meanwhile, empty GFP vector was used as a control. Resultantly, the vast majority of the green fluorescent signal of the TCD10-GFP fusion protein was coincident with the chlorophyll autofluorescence, and the remaining little parts did not overlap in tobacco cells (Fig. 8b). In contrast, the cells transformed with the empty GFP vector without a specific targeting sequence had green fluorescent signals in both the cytoplasm and the nucleus (Fig. 8a). These findings clearly suggest that TCD10 was mainly localized to the chloroplast in addition to secretory channels.
Discussion
TCD10 is Needed for Chloroplast Development and Growth under Cold Stress
The chloroplast is a semi-autonomous organelle, which contains about 100 genes, more than 3000 proteins function within it (Leister 2003). The normal chloroplast development needs tightly coordinated expressions between plastid- and nuclear genes. The mutation of those genes could result in chlorophyll deficient/chloroplast defects in plants. In this study, the tcd10 mutants, under low temperatures (20 °C), had the severe affected transcript-levels of most genes for Chl biosynthesis, photosynthesis and chloroplast development (Fig. 6), whereas the down-regulated genes at low temperatures could be recovered to normal level or even higher WT levels at high temperature (32 °C) (Fig. 7). This discrepancy was attributable to the difference in the chloroplast structure and pigment contents between 20 °C and 32 °C. In addition, as known, when rice seedlings grow up to the 3/4-leaf stage, a seed’s energy is exhausted, stored reserves become insufficient to meet itself the rice growth demand. The continued growing of seedlings requires the external supply of resources from photosynthesis. Hence, we could speculate that the no survival for tcd10 mutants after the 5-leaf stage at 20 °C maybe result from the loss of photosynthesis due to the block of electron transport in photosystems under cold stress. Notwithstanding the reason why abnormal chloroplast occurs only under cold stress is not completely claimed yet, it is speculated that the TCD10 function is possibly not prerequisite at higher temperatures, but it is essential/more required for rice chloroplast development under cold stress.
TCD10 Functions at the First Phase of Chloroplast Development Possible via Plastid Division
As described previously, the processes accompanying chloroplast development can be divided into three phases (Mullet 1993; Kusumi et al. 2010 and 2014). It is clearly known that OsPOLP1, encoding one plastidial DNA polymerase (Takeuchi et al. 2007; Kusumi et al. 2010), FtsZ encoding a component of the plastid division machinery (Vitha et al. 2001) are critical for the first phase, determine the number and size of chloroplasts (Takeuchi et al. 2007) and the NEP-transcribed genes which encode PEP cores (e.g. rpoA, rpoB, rpoC) for chloroplast genetic system and the PEP-transcribed genes (e.g. psab, LhcpII) involving PSII function at the second and third phase, respectively. Our data that all transcripts for the PEP-transcribed plastid (e.g. rbcL, psaA and LhcpII) and nuclear genes (rbcS) associated with photosynthesis apparatus (Fig. 6b) were obviously down-regulated in the tcd10 mutants showed that TCD10, at least, regulates the third phase of chloroplast development. In addition to V1 for NUS1 protein (Kusumi et al. 1997 and 2011), V2 for guanylate kinase (Sugimoto et al. 2007) and OsV4 for PPR protein (Gong et al. 2014) and ASL2 encoding plastid ribosomal protein L21 (Lin et al. 2015a, b) and TCM5 for Deg protease protein (Zheng et al. 2016), the extremely decreased accumulation of OsRpoTp (Hiratsuka et al. 1989) and the NEP-transcribed gene rpoB and rpoC which encodes PEP core β, β’ subunits functioning in the second step suggests that TCD10 also affects the second phase of chloroplast development. Similarly, the significantly reduced transcripts of rbcL, an indicator for the accumulation of polysomes (Barkan 1993), chloroplast 16SrRNA, 23SrRNA and rps7, rps14, rps20 in the tcd10 mutants indicated that the tcd10 mutation affects the assembly and accumulation of plastid ribosomes, resulting in disruption of plastid translation. Similarly, the blockage of the chloroplast 16SrRNA maturation in Arabidopsis rap mutant (Kleinknecht et al. 2014) and rice al1 mutant (Zhang et al. 2016) leads to abnormal chloroplasts. Thus, TCD10 participates in not only the plastid translation, but also the assembly and the accumulation of plastid ribosomes. More notably, the obviously down-regulations in transcripts of the known key genes (OsPoLP1 for one plastidial DNApolymerase (Takeuchi et al. 2007) (>25 fold) and RNRL for (Yoo et al. 2009) (>11 fold), OsDG2 for glycine-rich protein (Jiang et al. 2014a) (>166 fold) at the first step of chloroplast development indicated that TCD10 functions in the first step. As for the slightly increase in FtsZ transcript (<3 fold), it is likely due to the feedback effects. In addition, in view of fewer and smaller of chloroplasts (Fig. 2g) and the abnormal thylakoid structure (Fig. 2h) in tcd10 mutant cells, it suggests that the TCD10 regulates the first phase of chloroplast development probably via plasmid division under cold stress.
Possible Role of TCD10 Protein in Chloroplast RNA Metabolism
PPR protein can specifically identify single stranded RNA sequence, and it is found that the PPR protein is closely related to the transcription process, the shearing process, the editing process and the stability of RNA (Kotera et al. 2005; Okuda et al. 2007; Chateigner-Boutin et al. 2008; Cai et al. 2009; Yu et al. 2009; Zhou et al. 2009; Tseng et al. 2010; Sosso et al. 2012; Williams and Barkan 2003; Tavares-Carreón et al. 2008; Yamazaki et al. 2004; Pfalz et al. 2009; Fujii et al. 2013). With the unveiling of the primary version of the PPR code, manipulation of PPRs to bind to target RNAs is within reach (Barkan et al. 2012), biochemical (Barkan et al. 2012; Kobayashi et al. 2012), statistical (Barkan et al. 2012; Yagi et al. 2013), and evolutionary constraint (Fujii and Small 2011) studies have postulated that the 1st, 3rd, and 6th amino acids within the PPR consensus manipulates RNA base recognition specificity. As stated above, at low temperatures, TCD10 directly or indirectly regulates RNA transcript levels at all three steps of chloroplast development and participates in the plastid translation, the assembly and the accumulation of plastid ribosomes. As for TCD10, how to specifically participate in chloroplast RNA metabolism is unclear yet in detail. However, interestingly, Arabidopsis PGR3 (At4g31850), containing the same 27 PPR motifs, with the highest homolog (62.3% of identity) to TCD10 (Fig. 4a and Additional file 7: Figure S5), was previously studied in very detail (Shikanai et al. 1999; Yamazaki et al. 2004; Cai et al. 2011; Fujii et al. 2013) by mean of three Arabidopsis pgr3-1(the substitution mutation site in the 15th PPR motif), pgr3-2(the substitution mutation in the 12th PPR motif), and pgr3-3 (the substitution mutation in the 27th PPR motif) mutant alleles (Additional file 7: Figure S5). The chloroplast-targeted PGR3 was reported to have three distinct functions: (i) stabilization of photosynthetic electron transport L (petL) operon RNA, (ii) translational activation of petL, and (iii) translational activation of ndhA (Yamazaki et al. 2004; Cai et al. 2011) through binding to the 5′untranslated region (UTR) of petL operon RNA and ndhA (Cai et al. 2011). More interestingly, the 16 N-terminal PPRs in PRG3 was sufficient for function (i) via sequence-specific RNA binding, whereas the 11 C-terminal motifs were essential for functions (ii) and (iii) by activating translation (Fujii et al. 2013). Considering the high homolog between TCD10 and PGR3, it could be speculated that TCD10 may have similar three functions. Since the mutation in tcd10 mutants occurred in the 5th PPR motif site (Fig. 3e), resulting in the changes of structure and loss-of-function of TCD10, thus the tcd10 mutants should loss all three functions aforementioned. Indeed, the severe defects in photosynthetic electron transport pathways in tcd10 mutants as pgr3 mutants (Yamazaki et al. 2004) strongly supported the notion that TCD10 has the stabilization (i) of photosynthetic electron transport under cold stress; Meanwhile, the reduced transcripts of protochlorophyllide oxidoreductase also indirectly indicates the block of (iii) translational activation of ndhA. Regrettably, no evidences in our study can support the (ii) function of translational activation of petL because of our technical limitations. It is notable that, only under cold stress, TCD10 affected chloroplast development unlike Arabidopsis PRG3 without temperatures, showing their obviously different responses to temperatures. These findings could highlight the notion that highly conserved genes within various species might play diverse and complex roles than previously recognized. Hence, further understanding function difference between PGR3 and TCD10 and how TCD10 functions chloroplast development under cold stress is needed.
Conclusions
The rice TCD10 encodes a new PPR protein, mainly located in chloroplast, which is important for chloroplast development, seedling growth and the maintenance of photosynthetic electron transport and its disorder would lead to an aberrant chloroplast and abnormal transcript levels of genes associated with chloroplast development and photosynthesis under cold stress.
Methods
Plant Materials and Growth Conditions
The thermo-sensitive chlorophyll-deficient mutant, tcd10, used in this study, was isolated from japonica rice variety, Xiushui11 (WT), treated by 60Co gamma-radiation in 2006. All rice plants were grown in a rice paddy field in Shanghai (31°11’N, summer season, temperate climate) and Hainan (18°16’N, winter season, subtropical climate), China under local conditions or in the growth chambers.
Phenotype Observation and Photosynthetic Pigment Measurements
The germinated rice-seeds were grown at the chambers with four temperatures (20 °C, 24 °C, 28 °C and 32 °C) and 12-h-dark/12-h-light. Each third-leaf 200 mg were sampled at the 3-leaf stage and used to extract the pigments. Photosynthetic pigment (chlorophyll a, b carotenoid) were measured according to previously described methods (Jiang et al. 2014b).
To examine the changes of plant height and leaf chlorophyll content during the whole growth-period, leaf chlorophyll SPAD values, using fast non-destructive chlorophyll meter (SPAD-502, Minolta, Japan) (Peng et al. 1993; Turner and Jund, 1991), and plant height were measured once a week from the 1st weeks after transplanting, respectively. At maturity, panicle-associated traits of rice plants (Additional file 1: Figure S1) were investigated.
Chlorophyll Fluorescence and Transmission Electron Microscopy Analysis
Rice seedlings were cultivated at 20 °C and 32 °C in growth chambers. Chlorophyll fluorescence analysis for the 3rd leaves at the 3-leaf stage was performed by mean of a PAM-2000 portable chlorophyll fluorometer (MINI-PAM; Walz; http://walz.com). In addition to the max rate of photosynthetic electron transport (rETRmax), the variables Fo, Fm, Fv, and the Fv/Fm ratio were measured and calculated basically according to Meurer et al. (1996) after dark-adapted for 20 min. In addition, the observation of chloroplast ultrastructure in the third leaves was performed according to the previously described methods (Jiang et al. 2014a, b). Finally, samples were examined with a Hitachi-7650 transmission electron microscope.
Map-based Cloning of TCD10
Rice genomic DNA was extracted from young leaves by the CTAB method with minor modifications (Stewart and Via 1993). For genetic analysis and mapping for TCD10 locus, we crossed tcd10 mutant with Guangzhan63S (indica) to obtain F2 mapping population. Initially, we adopted 81 SSR primers based on data in Gramene (http://www.gramene.org) to determine the chromosome location of TCD10. Then new SSR and InDel markers (Additional file 5: Table S2) were designed using the PREMIRE 5.0 software based on the whole genomic sequences of the japonica Nipponbare variety (Goff et al. 2002) and the indica variety 9311 (Yu et al. 2002). In this study, a total of 5200 F2 mutant individuals were selected for fine-mapping. The functions of candidate genes and respective cDNA sequences were obtaining using TIGR (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/) and KOME (http://cdna01.dna.affrc.go.jp/cDNA/index.html), respectively.
RNAi Suppression and Knockout of TCD10
To confirm that TCD10 was blame for the mutant phenotype observed, both RNAi and CRISPR/Cas9 genome editing technology, which is of significance for basic plant research and crop genetic improvement (Belhaj et al. 2015; Ma and Liu 2016; Ma et al. 2016), were preformed. In RNAi experiments, the construct vector pTCK303 with a maize ubiquitin promoter and a rice intron was used as an RNAi vector (Wang et al. 2004). Both anti-sense and sense versions of a specific 780 bp fragment from the coding region of the TCD10 were amplified, and successively inserted into pTCK303, to form the RNAi construct vector pTCK303-dsRNAiTCD10. The primer pairs are 5'CGAGCTCGGTTCCTGATGTTCTTGCCGTG3' (SacI), 5' GACTAGTC GATCAGGTATGCATCCTTTCG 3' (SpeI) and 5' CGGGATCCCG GTTCCTGATGTTCTTGCCGTG 3 '(BamHI), 5' GGGGTACCCC GATCAGGTATGCATCCTTTCG 3' (KpnI).
In CRISPR/Cas9 system experiments, to generate the Cas9 targeting construct for TCD10, using the CRISPR Primer Deigner software (Naito et al. 2015; http://crispr.dbcls.jp/), the annealed gRNA oligonucleotide pair (F: 5'GCCGCGCGGCCATGGCCGGGTCG3'; R: 5'AAAC CGACCCGGCCATGGCCGCG3') with recognition sequence was designed. Then, the recognition sequence was inserted into the region between the OsU6 promoter and the gRNA scaffolds, from pYLgRNA-OsU6 vector, of Cas9 expression backbone vector (pYLCRISPR/Cas9-MH) at the BsaI sites according to the previously described method (Ma et al. 2015).
The resultant plasmid (pTCK303-dsRNAiTCD10 and CRISPR/Cas9 expression) and the respective empty vectors were introduced into Agrobacterium tumefaciens EHA105 and used to infect calli of WT plants according to the previously published methods (Hiei et al. 1994). The obtained T0 transgenic seedlings were grown in a paddy rice field after screening of tolerant-hygromycin and DNA sequencing according to our described methods (Jiang et al. 2014a, b). Then all T1 seedlings grown at 20 °C were used for surveying the segregation of phenotype.
RNA Transcript Analysis
Total RNA was extracted from various fresh tissues by the TRIzol Reagent (Invitrogen; http://www.invitrogen.com) and DNase I treated by an RNeasy kit (Qiagen; http://www.qiagen.com) and the first-strand cDNA was synthesized with the Revert-Aid first-strand cDNA synthesis kit (Toyobo; http://www.toyobo.co.jp) following the manufacturer’s instructions. The specific primers used for RT-PCR assays were shown in Additional file 9: Table S3. Then semiquantitative RT-PCR analysis was performed to assay the tissue expression pattern of TCD10. Real Time qPCR (ABI7500, Applied Biosystems; http://www.appliedbiosystems.com) was used to analyze RNA expression levels for associated-genes of chloroplast development, chlorophyll synthesis and photosynthesis. The relative quantification of gene expression data was analyzed as described by Livak and Schmittgen (2001). Three independent experiments were performed, and each sample was run in triplicate. OsActin was used as an internal control.
TCD10 Subcellular Localization
To investigate the subcellular localization of TCD10, cDNA fragments encoding the N-terminal region of 260 amino acids in TCD10 were amplified by PCR using following primer (5'-GAAGATCTATGTTGGAGGTCTGCTGCTGC-3' and 5'-GGGGTACCCCCTTCATTCTCCATTTTCGC-3') and introduced into vector pMON530-GFP at the BglII and KpnI sites (the sequence underlined were represented cleavage sites). TCD10 localization was investigated by transient expression of GFP fusion in tobacco (Nicotiana tabacum) cells, and confocal microscopy as described previously (Jiang et al. 2014a, b). Finally, GFP fluorescence in the transformed protoplasts was imaged using a confocal laser-scanning microscopy (LSM 5 PASCAL; ZEISS, http://www.zeiss.com)
Sequence and Phylogenetic Analysis
Gene prediction and structure analysis were performed using the GRAMENE database (www.gramene.org/). Homologous sequences of TCD10 were identified using the Blastp search program of the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov/). A phylogenetic tree of TCD10 and related proteins was then constructed by MEGA4 tool using the Neighbor-joining method with a bootstrap value of 1000 (Edgar 2004). The signal peptide was predicted with SignalP version 2.0 (Nielsen and Krogh 1998).
References
Barkan A (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5:389–402
Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS (2012) A combinatorial amino acid code forRNA recognition by pentatricopeptide repeat proteins. Plos Genet 8:263
Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V (2015) Editing plant genomes with crispr/cas9. Current Opinion in Biotechnology 32:76–84.
Cai W, Ji D, Peng L, Guo J, Ma J, Zou M, Lu C, Zhang L (2009) LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol 150:1260–1271
Cai W, Okuda K, Peng L, Shikanai T (2011), Proton gradient regulation 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J 67:318–327.
Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz G-NM, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, León P (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56:590–602
Chory J, Nagpal P, Peto CA (1991) Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3:445–459
De Longevialle AF, Meyer EH, Andrés C, Taylor NL, Lurin C, Millar AH, Small I (2007) The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19:3256–3265
De Santis-Maciossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rudiger W, Koop HU, Herrmann RG (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1. Molecular biology, biochemistry and ultrastructure. Plant J 18:477–489
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797.
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Fujii S, Small I (2011) The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol 191:37–47
Fujii S, Sato N, Shikanai T (2013) Mutagenesis of individual pentatricopeptide repeat motifs affects RNA binding activity and reveals functional partitioning of arabidopsis proton gradient regulation3. Plant Cell 25:3079–3088
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Gong X, Su Q, Lin D, Xu J, Jiang Q, Zhang J, Teng S, Dong Y (2014) The rice OsV4 encoding a novel pentatricopeptide repeat protein is required for chloroplast development during the early leaf stage under cold stress. J Integr Plant Biol 56:400–410
Gothandam KM, Kim ES, Cho H, Chung YY (2005) OsPPR1 a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol Biol 58:421–433
Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16:4041–4048
Hattori M, Miyake H, Sugita M (2007) A pentatricopeptide repeat protein is required for RNA processing of clpP Pre-mRNA in moss chloroplasts. J Biol Chem 282:10773–10782
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M (1989) The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet 217:185–194
Ichinose M, Tasaki E, Sugita C, Sugita M (2012) A PPR-DYW protein is required for splicing of a group II intron of cox1 premRNA in Physcomitrella patens. Plant J 70:271–278
Jiang Q, Ma X, Gong X, Zhang J, Teng S, Xu J, Lin D, Dong Y (2014a) The rice OsDG2 encoding a glycine-rich protein is involved in the regulation of chloroplast development during early seedling stage. Plant Cell Rep 33:733–744
Jiang Q, Mei J, Gong X, Xu J, Zhang J, Teng S, Lin D, Dong Y (2014b) Importance of the rice TCD9 encoding α subunit of chaperonin protein60 (Cpn60α) for the chloroplast development during the early leaf stage. Plant Sci 215–216:172–179
Kleinknecht L, Wang F, Stübe R, Philippar K, Nickelsen J, Bohne AV (2014) RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell 26(2):777–787
Kobayashi K, Kawabata M, Hisano K, Kazama T, Matsuoka K, Sugita M (2012) Identification and characterization of the RNA binding surface of the pentatricopeptide repeat protein. Nucleic Acids Res 40:2712–2723
Kocábek T, Řepková J, Dudová M, Hoyerová K, Vrba L (2005) Isolation and characterization of a novel semi-lethal Arabidopsis thaliana, mutant of gene for pentatricopeptide (PPR) repeat-containing protein. Genetica 128:395–407
Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433:326–330
Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Physiol 26:125–134
Kusumi K, Iba K (2014) Establishment of the chloroplast genetic system in rice during early leaf development and at low temperatures. Front Plant Sci 5:1–6
Kusumi K, Mizutani A, Nishimura M, Iba K (1997) A Virescent gene v1 determines the expression timing of plastid genes for transcription translation apparatus during early leaf development in rice. Plant J 12:1241–1250
Kusumi K, Chono Y, Gotoh E, Tsuyama M, Iba K (2010) Chloroplast biogenesis during the early stage of leaf development in rice. Plant Biotechnol 27:85–90
Kusumi K, Sakata C, Nakamura T, Kawasaki S, Yoshimura A, Iba K (2011) A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J 68:1039–1050.
Leister D (2003) Chloroplast research in the genomic age. Trends Genet 19:47–56
Lin D, Jiang Q, Zheng K, Chen S, Zhou H, Gong X, Xu J, Teng S, Dong Y (2015a) Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol 17(3):599–607
Lin D, Gong X, Jiang Q, Zheng K, Zhou H, Xu J, Teng S, Dong Y (2015b) The rice ALS3 encoding a novel pentatricopeptide repeat protein is required for chloroplast development and seedling growth. Rice 8:17
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Ma X, Liu Y-G (2016) CRISPR/Cas9-based multiplex genome editing in monocot and dicot plants. Curr Protoc Mol Biol 115:31.6.1
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein, involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15:1480–1495
Meurer J, Meierhoff K, Westhoff P (1996) Isolation of high-chlorophyll fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198:385–396.
Mullet JE (1993) Dynamic regulation of chloroplast transcription. Plant Physiol 103:309–313
Naito Y, Hino K, Bono H and Ui-Tei K (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31(7):1120–1123.
Nakamura T, Yagi Y, Kobayashi K (2012) Mechanistic insight into pentatricopeptide repeat proteins as sequence-specific RNA-binding proteins for organellar RNAs In plants. Plant Cell Physiol 53:1171–1179
Nielsen H, and Krogh A. (1998). Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol 6;122–130
Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T (2007) Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci U S A 104:8178–8183
Peng S, García FV, Laza RC, Cassman KG (1993) Adjustment for specific leaf weight improves chlorophyll meter's estimate of rice leaf nitrogen concentration. Agron J 85:987–990
Pfalz J, Bayraktar OA, Prikryl J, Barkan A (2009) Site-specific binding of a PPR protein defines and stabilizes 5’ and 3’ mRNA termini in chloroplasts. EMBO J 28:2042–2052
Schmitz-Linneweber C, Small I (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13:663–670
Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18:2650–2663
Shikanai T, Munekage Y, Shimizu K, Endo T, Hashimoto T (1999) Identification and characterization of Arabidopsis mutants with reduced quenching of chlorophyll fluorescence. Plant Cell Physiol 40:1134–1142
Small ID, Peeters N (2000) The PPR motif- a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25:46–47
Sosso D, Mbelo S, Vernoud V, Gendrot G, Dedieu A, Chambrier P, Dauzat M, Heurtevin L, Guyon V, Takenaka M, Rogowsky PM (2012) PPR2263, a DYW-Subgroup Pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 24:676–691
Stewart CN, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14:748–750
Su N, Hu ML, Wu DX, Wu FQ, Fei GL (2012) Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol 159:227–238
Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Jba K (2007) The rice nuclear gene, VTRESCENT 2, is essential for chloroplast development and encode a novel type of guanylate kinase targeted to plastids and mltochondra. Plant J 52:512–527
Takeuchi R, Kimura S, Saotome A, Sakaguchi K (2007) Biochemical properties of a plastidial DNA polymerase of rice. Plant Mol Biol 64:601–611
Tavares-Carreón F, Camacho-Villasana Y, Zamudio-Ochoa A, Shingú-Vázquez M, Torres-Larios A, Pérez-Martínez X (2008) The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J Biol Chem 283:1472–1479
Tseng CC, Sung TY, Li YC, Hsu SJ, Lin CL, Hsieh MH (2010) Editing of accD and ndhF chloroplast transcripts is partially affected in the Arabidopsis vanilla cream1 mutant. Plant Mol Biol 73:309–323
Turner FT, Jund MF (1991) Chlorophyll meter to predict nitrogen topdress requirement for semidwarf rice. Agron J 83:926–928.
Vitha S, Mcandrew RS, Osteryoung KW (2001) FtsZ ring formation at the chloroplast division site in plants. J Cell Biol 153:111–120.
Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Report 22:409–417
Williams PM, Barkan A (2003) A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J 36:675–686
Yagi Y, Hayashi S, Kobayashi K, Hirayama T, Nakamura T (2013) Elucidation of the rna recognition code for pentatricopeptide repeat proteins involved in organelle rna editing in plants. Plos ONE 8:42–43
Yamazaki H, Tasaka M, Shikanai T (2004) PPR motifs of the nucleusencoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J 38:152–163
Yoo SC, Cho SH, Sugimoto H, Li J, Kusumi K, Koh HJ, Iba K, Paek NC (2009) Rice Virescent3 and Stripel encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol 150:388–401
Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92
Yu QB, Jiang Y, Chong K, Yang ZN (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59:1011–1023
Zhang Z, Tan J, Shi Z, Xie Q, Xing Y, Liu C et al (2016) Albino leaf 1 that encodes the sole octotricopeptide repeat protein is responsible for chloroplast development in rice. Plant Physiol 171:1182–1191
Zheng K, Zhao J, Lin D, Chen J, Xu J, Zhou H. (2016). The ricetcm5gene encoding a novel deg protease protein is essential for chloroplast development under high temperatures: Rice 9(1),1–13.
Zhou W, Cheng Y, Yap A, Chateigner-Boutin AL, Delannoy E, Hammani K, Small I, Huang J (2009) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J 58:82–96
Acknowledgments
We sincerely thank Dr. Youbin Xiang (Stowers Institute for Medical Research, USA) for her critical reading of and suggestions for our manuscript. The authors are grateful to Prof. Zhongnan Yang (Shanghai Normal University) and Yaoguang Liu (State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University) for kindly providing pMON530-GFP vector, and the pYLgRNA-OsU6 and pYLCRISPR/Cas9-MH) vectors, respectively. This work was financially supported by Natural Science Foundation of China (No. 30971552) and Shanghai Municipal Education Commission of China (No.14YZ076, No.J50401), Shanghai Municipal Science and Technology Commission of China (12ZR1422000,16ZR1425300,16391900700), and the Science Program of Shanghai Normal University (SK20159).
Authors’ contributions
DL and YD provided the mutant seeds and generated F2 seeds for genotyping and phenotyping. LW, JW, XG and YL performed the experiments of phenotype assays and molecular analysis. DL, YD, and JX designed and discussed the research. LW, DL, YD wrote the manuscript. All authors approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1:
Figure S1. Changes of leaf chlorophyll SPAD from transplanting to maturity. (2010, Shanghai, China). (DOC 43 kb)
Additional file 2:
Figure S2. Changes of plant height from transplanting to maturity. (2010, Shanghai, China). (DOC 47 kb)
Additional file 3:
Figure S3. Comparison of panicle-related traits between tcd10 mutant and wild type (2010, Shanghai, China); PN, Panicle number; PL, Panicle length (cm); GW, 1000-grain weight (g). (DOC 38 kb)
Additional file 4:
Table S1. Genetic segregation of tcd10 mutants in the F2 population. (DOC 30 kb)
Additional file 5:
Table S2. The PCR-based molecular markers designed for fine mapping. (DOCX 14 kb)
Additional file 6:
Figure S4. Full-length cDNA sequence of TCD10 and mutation sites; The nucleotides with the blue (A), red (G) and green (CCATGGCCGGGTCGGGG) letters represent the deleted nucleotides in tcd10, T1-1 and T1-2 transgenic lines using CRISPR/Cas9 system technique; The sequences in the box represents the recognition sequences in CRIPPER/Cas9 experiments. (DOCX 21 kb)
Additional file 7:
Figure S5. The protein sequences of TCD10 and PGR3 (At4g31850); The sequences with green letters indicate chloroplast transit peptide (CTP); The numbers represent PPR motifs; In TCD10, the amino acid with red letter indicate the deletion mutation sites in tcd10 (the 1st PPR) and T1-1, T1-2 transgenic lines (the 5th PPR), respectively; In PGR3, the amino acid with red letter indicate the substitute mutation sites in pgr3-1(the 15th PPR), pgr3-2(the 12th PPR) and pgr3-3(the 27th PPR) mutants. (DOCX 21 kb)
Additional file 8:
Figure S6. Expression patterns of TCD10 (LOC_Os10g28600). Data were obtained from the rice expression profile database, RiceXPro (http://ricexpro.dna.affrc.go.jp/category-select.php). (DOC 98 kb)
Additional file 9:
Table S3. Markers designed for realtime RT-PCR and gene functions. (DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wu, L., Wu, J., Liu, Y. et al. The Rice Pentatricopeptide Repeat Gene TCD10 is Needed for Chloroplast Development under Cold Stress. Rice 9, 67 (2016). https://doi.org/10.1186/s12284-016-0134-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-016-0134-1