Abstract
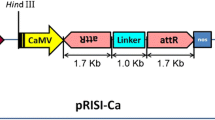
In the last decade, RNA interferences (RNAi) has proven to be an effective strategy to knock out homologous genes in a wide range of species. Based on its principle, a new generation of vectors containing an inverted target sequence separated by an intron as a loop, developing simplifications to the procedure of RNAi construction are required to improve the efficiency of gene inactivation techniques. Here, a novel polymerase chain reaction (PCR)—based RNAi vector pTCK303 with a maize ubiquitin promoter, 2 specific multiple enzyme sites, and a rice intron was constructed for monocot gene silencing. With this vector, only 1 PCR product amplified by a single pair of primers and 2 ligation reactions were needed to create an RNAi construct, which shortened the time span before being transformed into the plant. To test the efficiency of vector pTCK303, a rice geneOsGAS1 was used, and its RNAi construct was introduced into rice calli. Southern blot analysis of the transgenic rice confirmed the presence of theOsGAS1 RNAi structure. The decrease inOsGAS1 level in the transgenic rice was detected by Northern blot probed with anOsGAS1-specific sequence. Moreover, the rate of inhibition of the RNA expression level in RNAi transgenic rice was approximately 85% according to our real-time PCR. Therefore, the RNAi vector pTCK303 based on the homology-dependent gene-silencing mechanisms facilitated the inhibition of endogenous genes in a monocot and was proven to be a practical and efficient platform for silencing a rice gene.
Similar content being viewed by others
Abbreviations
- CaMV:
-
cauliflower mosaic virus
- DDCT method:
-
the comparative cycle threshold (CT) method
- dsRNA:
-
double-stranded RNA
- GUS:
-
β-glucuronidase
- MCS:
-
multiply clone site
- miRNAs:
-
microRNAs
- mRNA:
-
messenger RNA
- MS medium:
-
Murashige and Skoog medium
- NOS:
-
nopaline synthase
- PCR:
-
polymerase chain reaction
- RNAi:
-
RNA interference
- siRNAs:
-
small interfering RNAs
- X-Gluc:
-
5-bromo-4-chloro-3-indolyl-β-d-GlcUA
References
Christensen AH and Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5: 213–218.
Chuang CF and Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA inArabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985–4990.
Denli AM and Hannon GJ (2003) RNAi: an ever-growing puzzle. Trends Biochem Sci 28: 196–201.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, and Mello CC (1998) Potent and specific genetic interference by double-stranded RNA inCaenorhabditis elegans. Nature 391: 806–811.
Flavell RB (1994) Inactivation of gene expression in plants as a consequence of specific sequence duplication. Proc Natl Acad Sci USA 91: 3490–3496.
Ge L, Chen H, Jiang JF, Zhao Y, Xu ML, Xu YY, Tan KH, Xu ZH, and Chong K (2004) Overexpression ofOsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol 135: 1502–1513.
Guo S and Kemphues KJ (1995) par-1, a gene required for establishing polarity inC. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620.
Hunter CP (2000) Gene silencing: shrinking the black box of RNAi. Curr Biol 10: R137–140.
Jefferson RA (1989) The GUS reporter gene system. Nature 342: 837–838.
Karsai A, Muller S, Platz S, and Hauser MT (2002) Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 32: 790–796.
Klink VP and Wolniak SM (2000) The efficacy of RNAi in the study of the plant cytoskeleton. J Plant Growth Regul 19: 371–384.
Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, and Carthew RW (2004) Distinct roles forDrosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81.
Pham JW, Pellino JL, Lee YS, Carthew RW, and Sontheimer EJ (2004) A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi inDrosophila. Cell 117: 83–94.
Sambrook J and Russell DW (2001) Molecular cloning: a laboratory manual. 3rd edition. pp 532–552. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Schweizer P, Pokorny J, Schulze-Lefert P, and Dudler R (2000) Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J 24: 895–903.
Svoboda P, Stein P, Hayashi H, and Schultz RM (2000) Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127: 4147–4156.
Tabara H, Grishok A, and Mello CC (1998) RNAi inC. elegans: soaking in the genome sequence. Science 282: 430–431.
Tijsterman M and Plasterk RH (2004) Dicers at RISC; the mechanism of RNAi. Cell 117: 1–3.
Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, and others (2001) Construct design for efficient, effective, and high-throughput gene silencing in plants. Plant J 27: 581–590.
Yoshida A, Suzuki N, Nakano Y, Oho T, Kawada M, and Koga T (2003) Development of a 5′ fluorogenic nuclease-based real-time PCR assay for quantitative detection ofActinobacillus actinomycetemcomitans andPorphyromonas gingivalis. J Clin Microbiol 41: 863–866.
Yoshinari K, Miyagishi M, and Taira K (2004) Effects on RNAi of the tight structure, sequence, and position of the targeted region. Nucleic Acids Res 32: 691–699.
Zamore PD (2001) RNA interference: listening to the sound of silence. Nat Struct Biol 8: 746–750.
Author information
Authors and Affiliations
Additional information
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wang, Z., Chen, C., Xu, Y. et al. A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22, 409–417 (2004). https://doi.org/10.1007/BF02772683
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF02772683