Abstract
Background
Abnormalities of chromosome 16 are found in about 5–8% of acute myeloid leukemia (AML). The AML with inv(16)(p13.1q22) or t (16;16)(p13.1;q22) is associated with a high rate of complete remission (CR) and favorable overall survival (OS) when treated with high-dose Cytarabine. At the inversion breakpoints, deletion of 3’CBFB has been reported, but most of them were studied by chromosome and fluorescence in situ hybridization (FISH) analyses. The genomic characteristics of such deletions remain largely undefined, hindering further understanding of the clinical significance of the deletions.
Case presentation
We report here two AML cases with inv(16) and deletion of the 5’MYH11/3’CBFB gene fusion, which were characterized by chromosome, FISH, and single nucleotide polymorphism (SNP) microarray analyses. Both cases have achieved CR for more than three years.
Conclusions
Deletion of 3’CBFB in AML with inv(16) is also accompanied with deletion of 5’MYH11 in all the cases studied by SNP microarray, suggesting that 3’CBFB and 5’MYH11 were most likely deleted together as a fusion product of inv(16) instead of occurring separately. In concert with the findings of other published studies of similar patients, our study suggests that deletion of 5’MYH11/3’CBFB in AML with inv(16) may not have negative impact on the prognosis of the disease.
Similar content being viewed by others
Background
Abnormalities of chromosome 16 are found in about 5–8% of acute myeloid leukemia (AML) and are one of the three AML defining chromosomal aberrations regardless of blast percentage under the World Health Organization (WHO) classification [1]. The AML with inv(16)(p13.1q22) or t (16;16)(p13.1;q22) is associated with a high rate of complete remission (CR) and favorable overall survival (OS) when treated with high-dose Cytarabine [2, 3]. The inv(16) results in a leukemogenic CBFB/MYH11 gene fusion [4, 5]. However, additional chromosome changes and/or gene mutations, such as + 22, + 8, deletion of 7q, and the CBL, FLT3, KIT gene mutations, are frequently found in AML with inv(16). These additional changes/mutations may influence the OS positively or negatively [6, 7]. For example, gain of an additional chromosome 22 in AML with inv(16) may predict an improved outcome [6, 8], whereas KIT mutations appear to have an increased risk of relapse and shorter survival in adult patients [6, 9]. Less commonly, deletions, especially deletion of 3’CBFB, may occur at the inversion breakpoints. Fourteen cases with inv(16) and deletion of 3’CBFB have been reported in literature [10,11,12,13,14,15,16,17,18,19]. However, only three of them were studied by microarray analysis, and only one was reported with unambiguous breakpoint coordinates [10]. The genomic characteristics of the vast majority of the reported 3’CBFB deletions were undefined.
Here we reported two AML with inv(16) cases, both carried an additional deletion at one inversion breakpoint involving the fusion between 5’MYH11 at 16p13.1 and 3’CBFB at 16q22. The genomic characteristics of both cases were characterized by chromosome, FISH and SNP microarray analyses. We also reviewed similar cases reported in literature to investigate possible clinical significance of the deletions at the inv(16) breakpoints.
Case presentation
Case 1
A 24-year-old male presented with intermittent fever and sore throat. A complete blood examination demonstrated a hemoglobin (Hb) count of 70 g/L, a white blood cell (WBC) count of 170 × 109/L with 80% blasts and a platelet count of 25 × 109/L. He had no hepatosplenomegaly. His bone marrow (BM) aspirate exhibited greater than 90% myelomonoblastic cells, with increased maturing eosinophils. Cytochemical staining was positive for peroxidase and esterase. Flow cytometry showed 76% positive CD34 and 5% positive CD64, but negative CD14, consistent with a diagnosis of AML. The patient started induction chemotherapy and achieved a complete hematological recovery on day 21. He then received autologous stem cell transplant (ASCT) after two cycles of consolidation chemotherapy and has remained in CR for three years.
Case 2
A 47-year-old male presented with intermittent low-grade fever and progressive fatigue. A complete blood examination demonstrated a Hb count of 92 g/L, a WBC count of 3.5 × 109/L and a platelet count of 43 × 109/L. This patient had splenomegaly. His BM aspirate showed 50% myeloid blasts. Flow cytometry showed 50% blasts expressed CD34, CD117, CD13, CD33, HLA-DR, and myeloperoxidase (MPO), consistent with a diagnosis of AML. The patient started induction chemotherapy and achieved a complete hematological recovery. He then received two cycles of consolidation chemotherapy. The patient has been in CR for four years.
Methods
Chromosome and FISH analyses
Chromosome G-banding was performed following standard techniques on BM aspirate. FISH was carried out with the commercial CBFB dual-color break-apart probe kit (Abbott Laboratories, Lake Bluff, IL) following the manufacturer’s protocol. The results were analyzed using the Leica CytoVision system (Leica Biosystems, San Jose, CA).
SNP microarray analysis
Genomic DNA was extracted from BM using QIAGEN EZ1 kit (Qiagen, Hilden, Germany). SNP microarray was set up using Illumina CytoSNP-850 K v1.1 BeadChip (Illumina, San Diego, CA) and analyzed using BlueFuse Multi (Illumina), based on human genome build GRCh37/hg19.
The nomenclature for the chromosome, FISH and array findings is based on the International System for Human Cytogenomic Nomenclature (ISCN) 2016 [20].
Results
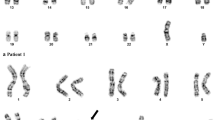
In case 1, chromosome analysis detected an abnormal karyotype with a pericentric inversion of chromosome 16 in all 20 cells analyzed (Fig. 1a), consistent with the diagnosis of AML with inv(16)(p13.1q22); CBFB-MYH11. In addition, 4 of these cells showed an extra copy of chromosome 22, which is a common secondary change in this disease. FISH analysis confirmed the inversion but also detected a deletion of 3’CBFB in approximately 95.5% of the interphase cells examined (Fig. 1a). SNP microarray confirmed the gain of chromosome 22 and a 1.1 Mb deletion involving 3’CBFB on the long arm of chromosome 16 within 16q22.1, and further detected a 416 Kb deletion involving the 5’MYH11 gene on the short arm of chromosome 16 within 16p13.11. The 16q22.1 deletion involved 53 known genes including 3’CBFB, and the 16p13.11 deletion involved 7 known genes including 5’MYH11 (Fig. 1b). The nomenclature of the cytogenomic findings in case 1 can be described as: 46,XY,inv(16)(p13.1q22)[16]/47,idem,+ 22[4].nuc ish (5’CBFBx2,3’CBFB×1)(5’CBFB con 3’CBFB×1)[191/200].arr[GRCh37] 16p13.11(15875744_16291983)×1[0.9],16q22.1(67128019_68214140)×1[0.9],(22) ×3[0.8].
a Chromosome 16 with inv(16)(p13.1q22), 3’CBFB deletion detected by interphase FISH (5’CBFB, red; 3’CBFB, green), and two small deletions on chromosome 16 detected by SNP microarray. b Genes in the deleted regions (black bars), adopted from UCSC genome browser (https://www.genome.ucsc.edu). Case 3 was from Dawson et al. [10], and COSMIC cancer genes were underlined
In case 2, a similar inversion of 16 with gain of additional chromosomes 9 and 22 was detected in all 20 metaphases analyzed by chromosome analysis (Fig. 1a). FISH revealed a similar abnormal signal pattern with a deletion of 3’CBFB in approximately 83.5% of the interphase cells examined (Fig. 1a). Similar to case 1, SNP microarray confirmed the chromosome and FISH findings and detected an additional deletion on the short arm of chromosome 16 (Fig. 1a). The 16p13.11p12.3 deletion is 1.1 Mb in size involving 14 known genes including 5’MYH11, and the 16q22.1 deletion is 986 kb in size involving 52 genes including 3’CBFB (Fig. 1b). The cytogenomic findings of case 2 can be described as: 48,XY,+ 9,inv(16)(p13.1q22),+ 22[20].nuc ish(5’CBFBx2,3’CBFB×1)(5’CBFB con 3’CBFB×1) [167/200].arr[GRCh37] (9)×3[0.7],16p13.11p12.3(15817490_16869754)×1[0.7],16q22.1(67113418_68099821)×1[0.7],(22)×3[0.7].
Discussion and conclusion
AML with inv(16) and deletion of 3’CBFB has been reported in at least 14 cases and most of the deletions were detected by FISH using CBFB break-apart probes [10,11,12,13,14,15,16,17,18,19]. To our knowledge, only three cases were analyzed using microarray, and of these cases, only one reported unambiguous genomic coordinates of the deletion [10, 11]. Noteworthily, all the cases analyzed with microarray, including the ones in this report, showed an additional deletion of 5’MYH11, suggesting that the 3’CBFB and 5’MYH11 were most likely deleted together as a fusion product of inv(16) instead of being deleted separately. More than 10 CBFB-MYH11 fusion transcripts with different sizes due to various genomic breakpoints have been reported [21, 22]. In this study, CBFB exons 1–5 fused with MYH11 exons 8–42 in case 1 and CBFB exons 1–4 fused with MYH11 exons 33–42 in case 2. Both are typical gene fusions in AML-associated inv(16), and the 5’CBFB/3’MYH11 fusion genes are believed to be responsible for the disease of the patients [23]. However, there is no evidence of additional phenotypical effects of deletion of the 5’MYH11/3’CBFB fusion genes. Both patients in this study responded to chemotherapy well and achieved CR for multiple years. Three cancer-related genes listed in the Cancer Gene Census (CGC) in the Catalogue of Somatic Mutations in Cancer (COSMIC) were involved in the deleted regions, including MYH11 in 16p, CBFB and CTCF in 16q (Fig. 1b) [24]. CBFB/MYH11 gene fusion caused by inv(16) or t (16;16) results in two fusion genes, 5’CBFB/3’MYH11 and 5’MYH11/3’CBFB. The former is a known pathogenic cause of AML, but the pathogenic effect of the latter is uncertain. We further reviewed reported AML cases with inv(16) and deletion of 3’CBFB (Table 1) and did not find significant phenotypic differences caused by the deletion of 3’CBFB. Of the 14 cases with clinical data available, 11 (79%) were known to achieve CR with known survival time up to 48 months at the time of report. This small cohort data appears to be in line with the CR rate (86–88%) and the five-year OS rate (50%) in AML with inv(16) [8, 25]. Taken together, the findings suggest that the 5’MYH11/3’CBFB fusion gene may play minimal roles in AML pathogenesis. CTCF is thought to be a tumor suppresser gene [26]. Kemp et al. reported CTCF haploinsufficiency destabilized DNA methylation and predisposed mice to cancer [27]. However, deletion of this gene in the current cases apparently did not result in additional phenotypic changes. Such changes, if there are any, may be less significant or overlapped with that of the AML with inv(16). Nevertheless, the significance of deletion of CTCF in AML with inv(16) needs to be further investigated. There are five cases with inv(16) and deletion of 3’CBFB, which either did not achieve CR or relapsed (Table 1). Since the deletion sizes and other genomic characteristics of these cases were undefined, it is unclear whether additional genes or other genomic elements were affected. Array analysis of similar cases may provide more insights on genomic changes and corresponding pathogenic effects in these cases.
Deletion of the 3’CBFB gene is a recurrent finding in AML with inv(16). It most likely represents deletions of the 5’MYH11/3’CBFB gene fusion from the inv(16) as a secondary change following the inversion. The 5’CBFB/3’MYH11 fusion resulting from the inv(16) is a known pathogenic cause of AML, but the 5’MYH11/3’CBFB fusion may play a minimal role in AML pathogenesis. The CTCF gene adjacent to the 3’CBFB was deleted in the current cases, suggesting that deletions of CTCF in AML with inv(16) may not have prognostic significance either. However, potential pathogenic effects of other genes involved in extended deletion regions may not be excluded.
Availability of data and materials
The data used or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AML:
-
Acute myeloid leukemia
- ASCT:
-
Autologous stem cell transplant
- BM:
-
Bone marrow
- CGC:
-
Cancer Gene Census
- COSMIC:
-
Catalogue of Somatic Mutations in Cancer
- CR:
-
Complete remission
- FISH:
-
Fluorescence in situ hybridization
- Hb:
-
Hemoglobin
- ISCN:
-
International System for Human Cytogenomic Nomenclature
- MPO:
-
Myeloperoxidase
- OS:
-
Overall survival
- SNP:
-
Single nucleotide polymorphism
- WBC:
-
White blood cell
References
Yang JJ, Park TS, Wan TS. Recurrent cytogenetic abnormalities in acute myeloid leukemia. Methods Mol Biol. 2017;1541:223–45.
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–65.
Harrison CJ, Hills RK, Moorman AV, Grimwade DJ, Hann I, Webb DK, et al. Cytogenetics of childhood acute myeloid leukemia: United Kingdom Medical Research Council treatment trials AML 10 and 12. J Clin Oncol. 2010;28(16):2674–81.
Castilla LH, Garrett L, Adya N, Orlic D, Dutra A, Anderson S, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23(2):144–6.
Kuo YH, Landrette SF, Heilman SA, Perrat PN, Garrett L, Liu PP, et al. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9(1):57–68.
Paschka P, Du J, Schlenk RF, Gaidzik VI, Bullinger L, Corbacioglu A, et al. Secondary genetic lesions in acute myeloid leukemia with inv(16) or t (16;16): a study of the German-Austrian AML study group (AMLSG). Blood. 2013;121(1):170–7.
Itzykson R, Duployez N, Fasan A, Decool G, Marceau-Renaut A, Meggendorfer M, et al. Clonal interference of signaling mutations worsens prognosis in core-binding factor acute myeloid leukemia. Blood. 2018;132(2):187–96.
Marcucci G, Mrozek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t (8;21) differ from those of patients with inv(16): a Cancer and leukemia group B study. J Clin Oncol. 2005;23(24):5705–17.
Paschka P, Marcucci G, Ruppert AS, Mrozek K, Chen H, Kittles RA, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t (8;21): a Cancer and leukemia group B study. J Clin Oncol. 2006;24(24):3904–11.
Dawson AJ, Bal S, McTavish B, Tomiuk M, Schroedter I, Ahsanuddin AN, et al. Inversion and deletion of 16q22 defined by array CGH, FISH, and RT-PCR in a patient with AML. Cancer Genet. 2011;204(6):344–7.
Haferlach C, Dicker F, Kohlmann A, Schindela S, Weiss T, Kern W, et al. AML with CBFB-MYH11 rearrangement demonstrate RAS pathway alterations in 92% of all cases including a high frequency of NF1 deletions. Leukemia. 2010;24(5):1065–9.
Tirado CA, Valdez F, Klesse L, Karandikar NJ, Uddin N, Arbini A, et al. Acute myeloid leukemia with inv(16) with CBFB-MYH11, 3'CBFB deletion, variant t (9;22) with BCR-ABL1, and del (7)(q22q32) in a pediatric patient: case report and literature review. Cancer Genet Cytogenet. 2010;200(1):54–9.
Spencer DV, Cavalier M, Kalpatthi R, Quigley DI. Inverted and deleted chromosome 16 with deletion of 3'CBFB identified by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2007;179(1):82–4.
Hung D, St Heaps L, Benson W, Mirochnik O, Sharma P, Smith A. Deletion of 3'CBFbeta in an inv(16)(p13.lq22) ascertained by fluorescence in situ hybridization and reverse-transcriptase polymerase chain reaction. Cancer Genet Cytogenet. 2007;172(1):92–4.
Kelly J, Foot NJ, Conneally E, Enright H, Humphreys M, Saunders K, et al. 3'CBFbeta deletion associated with inv(16) in acute myeloid leukemia. Cancer Genet Cytogenet. 2005;162(2):122–6.
Egan N, O'Reilly J, Chipper L, Higgins M, Herrmann R, Cannell P. Deletion of CBFB in a patient with acute myelomonocytic leukemia (AML M4Eo) and inversion 16. Cancer Genet Cytogenet. 2004;154(1):60–2.
Kolomietz E, Al-Maghrabi J, Brennan S, Karaskova J, Minkin S, Lipton J, et al. Primary chromosomal rearrangements of leukemia are frequently accompanied by extensive submicroscopic deletions and may lead to altered prognosis. Blood. 2001;97(11):3581–8.
Pirc-Danoewinata H, Dauwerse HG, Konig M, Chudoba I, Mitterbauer M, Jager U, et al. CBFB/MYH11 fusion in a patient with AML-M4Eo and cytogenetically normal chromosomes 16. Genes Chromosomes Cancer. 2000;29(2):186–91.
Batanian JR, Huang Y, Fallon R. Deletion of 3′-CBFB gene in association with an inversion (16)(p13q22) and a loss of the Y chromosome in a 2-year-old child with acute myelogenous leukemia-M4. Cancer Genet Cytogenet. 2000;121(2):216–9.
McGowan-Jordan J, Simons A, Schmid M, editors. ISCN 2016, An International System for Human Cytogenetic Nomenclature (2016). Basel: Karger Press; 2016.
Schwind S, Edwards CG, Nicolet D, Mrozek K, Maharry K, Wu YZ, et al. inv(16)/t (16;16) acute myeloid leukemia with non-type a CBFB-MYH11 fusions associate with distinct clinical and genetic features and lack KIT mutations. Blood. 2013;121(2):385–91.
Park TS, Lee ST, Song J, Lee KA, Lee JH, Kim J, et al. Detection of a novel CBFB/MYH11 variant fusion transcript (K-type) showing partial insertion of exon 6 of CBFB gene using two commercially available multiplex RT-PCR kits. Cancer Genet Cytogenet. 2009;189(2):87–92.
Liu PP, Hajra A, Wijmenga C, Collins FS. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood. 1995;85(9):2289–302.
Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–83.
Solh M, Yohe S, Weisdorf D, Ustun C. Core-binding factor acute myeloid leukemia: heterogeneity, monitoring, and therapy. Am J Hematol. 2014;89(12):1121–31.
Nichols MH, Corces VG. A CTCF code for 3D genome architecture. Cell. 2015;162(4):703–5.
Kemp CJ, Moore JM, Moser R, Bernard B, Teater M, Smith LE, et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7(4):1020–9.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
LL and ZQ collected the data and wrote the draft of the manuscript. JY reviewed the manuscript and made critical revision of the manuscript before submission. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All patient-related data were collected retrospectively and anonymously. The study was approved by the Institutional Review Board (IRB) of University of California San Francisco.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lv, L., Yu, J. & Qi, Z. Acute myeloid leukemia with inv(16)(p13.1q22) and deletion of the 5’MYH11/3’CBFB gene fusion: a report of two cases and literature review. Mol Cytogenet 13, 4 (2020). https://doi.org/10.1186/s13039-020-0474-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-020-0474-9