Abstract
Background
Camelina sativa (L.) Crantz, known by such popular names as “gold-of-pleasure” and “false flax,” is an alternative oilseed crop for biofuel production and can be grown in harsh environments. Considerable interest is now being given to the new concept of the development of a fusion plant which can be used as a soil remediation plant for ground contaminated by heavy metals as well as a bioenergy crop. However, knowledge of the transport processes for heavy metals across Camelina plant membranes is still rudimentary.
Results
Firstly, to investigate whether Camelina HMA (heavy metal P1B-ATPase) genes could be used in such a plant, we analyzed the expression patterns of eight HMA genes in Camelina (taken from the root, leaf, stem, flower, and silique). CsHMA3 genes were expressed in all organs. In addition, CsHMA3 was induced in roots and leaves especially after Pb treatment. Heterogeneous expression of CsHMA3 complemented the Pb- or Zn-sensitive phenotype of Δycf1 or Δzrc1 yeast mutant strains. Subsequently, we cloned and overexpressed CsHMA3 in Camelina. The root growth of transgenic lines was better than that in the wild-type plant under heavy metal stress (for Cd, Pb, and Zn). In particular, the transgenic lines showed enhanced Pb tolerance in a wide range of Pb concentrations. Furthermore, the Pb and Zn content in the shoots of the transgenic lines were higher than those in the wild-type plant. These results suggest that overexpression of CsHMA3 might enhance Pb and Zn tolerance and translocation. Also, the transgenic lines displayed a wider leaf shape compared with the wild-type plant due to an induction of genes related to leaf width growth and showed a greater total seed yield compared to the wild type under heavy metal stress.
Conclusions
Our data obtained from physiological and functional analyses using CsHMA3 overexpression plants will be useful to develop a multifunctional plant that can improve the productivity of a bioenergy crop and simultaneously be used to purify an area contaminated by various heavy metals.
Similar content being viewed by others
Background
Nowadays, the requirement for fossil fuels is increasing exponentially, and it is calculated that the remaining fossil fuel reserves will be consumed by the year 2050 [1–3]. Vegetable oils are widely recognized as promising alternatives to regular diesel fuel, because they are renewable and environmentally friendly. Among the several renewable energy sources, oilseed crops are increasingly being considered as materials to fulfill the rising demand of fossil fuels and their products [4]. Camelina sativa, known by its popular names of “gold-of-pleasure” and “false flax,” is an alternative oilseed crop that can be used as a potential low-input coast oil crop for biodiesel production and can be grown under different climatic and soil conditions. Although it was widely grown in Europe and Russia until the 1940s, Camelina was largely displaced by higher yielding crops after World War II [5–8].
In recent years, Camelina production has increased somewhat due to the heightened interest in vegetable oils that are high in omega-3 fatty acids (a principal component of Camelina oil) [9, 10]. Camelina has a short growing season (85 to 100 d), so it can be incorporated into double cropping systems during the period between harvest and planting times of the main crop. However, the full potential of this crop has not yet been explored, and very little plant breeding or crop production improvement has been performed on Camelina.
To meet the increasing demand for oilseed crops, the cultivation of oil crops on a large scale needs to be considered without wasting agricultural land. Therefore, other types of land which we consider as having barren soil can be used for cultivation following a suitable scientific approach.
Heavy metal contamination is a serious problem of increasing significance for environmental reasons. In particular, many crops are exposed to cadmium (Cd) and lead (Pb) around abandoned mines. Therefore, considerable interest has developed recently in the use of terrestrial plants as a green technology for the remediation of surface soils contaminated with toxic heavy metals [11, 12]. Interest in phytoremediation has grown significantly following the identification of metal hyperaccumulator plant species. Hyperaccumulators are conventionally defined as plants having a Zn concentration above 10,000 μg g-1 dw, a Pb and Co concentration higher than 1000 μg g-1 dw, or a Cd concentration above 100 μg g-1 dw [13, 14].
Based on genetic and functional studies of the Arabidopsis P1B-ATPases, it is predicted that certain pumps (HMA1 to 4) will be capable of transporting Cd, Pb, Zn, and Co, whereas other pumps (HMA 5 to 8) could transport Cu and potentially Ag [15–19]. Meanwhile, the presence of a multiple genomic copy number can enhance the expression levels of some candidate genes for metal hyperaccumulation and tolerance in hyperaccumulators [20–22]. According to a recent study, genes in Camelina were found in two or three copies, and Camelina has a hexaploid genome [23]. In this study, we investigated the productivity index between wild-type and CsHMA3 transgenic lines, and tested whether Camelina HMA3 (heavy metal P1B-ATPase 3) can enhance functions in metal accumulation or tolerance. The results obtained from the present study provide knowledge on how to improve the growth of a biodiesel species of Camelina on different concentrations of heavy metals, such as Cd, Pb, Zn, and Co.
Results
Effect of heavy metal stress on physiological responses in Camelinaand rapeseed
Rapeseed (Brassica napus L.) is another alternative oilseed crop and is known as a potentially useful candidate for phytoremediation [24, 25]. To study whether Camelina can also be used for phytoremediation, we performed a comparative analysis of the physiological responses of Camelina and rapeseed under heavy metal stress. Firstly, we have investigated the effect of metal in a concentration-dependent manner on root growth and germination. Metal-treated root was able to grow with up to 50 μM Cd, 500 μM Pb, 500 μM Zn, and 100 μM Co. Beyond this range, the root growth rate was drastically reduced (data not shown). This result indicates that the critical concentration for growth of both plants is considered to be 50 μM Cd, 500 μM Pb, 500 μM Zn, and 100 μM Co in our system. We also observed that root growth rate and electrolyte leakage were affected by heavy metal treatment in both plants; however, Camelina treated with Cd and Pb exhibited less inhibition of root growth rate, and leaves treated with Pb showed a lower amount of leakage than that of rapeseed (Figure 1; Additional file 1: Figure S1).
Effect of metal treatments on root growth in Camelina and rapeseed. Seeds were germinated and grown on agar plates containing 50 μM Cd, 500 μM Pb, 500 μM Zn, and 100 μM Co for seven days. Data represent means ± SD (n = 30). Mean values of root length in the normal medium as control for Cd, Zn, and Co were 4.10 ± 0.69 (Camelina) and 4.62 ± 0.53 (rapeseed); mean values of root length in the no-phosphate medium control for Pb were 3.25 ± 0.11 (Camelina) and 3.80 ± 0.55 (rapeseed). Asterisks in the figure indicate significant difference between Camelina and rapeseed subjected to the same treatment at ***P < 0.001 by Student’s t-test.
To compare the heavy metal uptake ability between Camelina and rapeseed, we measured the Cd, Pb, Zn, and Co content in Camelina and rapeseed grown in solution with either 50 μM Cd, 500 μM, 500 μM Zn, or 100 μM Co, respectively, for one week (Figure 2). Although the Cd content in roots of rapeseed seems slightly higher than that of Camelina, the difference was not statistically significant. Also, no statistical difference was shown in the Pb and Zn contents in roots between Camelina and rapeseed. The Co content in Camelina root was lower than that of rapeseed. In the shoots, the Cd, Pb, Zn, and Co contents in Camelina were higher than those of rapeseed (Figure 2). When comparing physiological parameters for long-term stress between Camelina and rapeseed, no statistical differences were observed in chlorophyll content, stomata conductance, and growth rate (data not shown). These results suggest that Camelina also has the potential to be used for phytoremediation along with rapeseed.
Metal contents in roots and shoots of Camelina and rapeseed .Four-week-old plants were grown hydroponically and treated with 50 μM Cd, 500 μM Pb, 500 μM Zn, and 100 μM Co for one week. Data represent means ± SD (n = 3) from three independent plants. Asterisks in the figure indicate significant difference between Camelina and rapeseed subjected to the same treatment at *P < 0.05, **P < 0.01, and ***P < 0.001 by Student’s t-test.
Identification and phylogenetic analysis of CsHMA3gene
Our next research task involved the possibility of a new transgenic plant that is better suited for heavy metal phytoremediation. To determine if Camelina genes could be used in such a plant, the full-length cDNA of Camelina sativa L. HMA3 (CsHMA3) was first isolated from cultivar “CAME” with 5’ and 3’ PCR primers. Based on the Camelina Genome Project Portal (http://www.Camelinagenomics.org and http://www.Camelinagenomics.net; non-public databases), we found that the CsHMA3 gene occurs in three copies in the genome of Camelina, named CsHMA3a, CsHMA3b, and CsHMA3c. The CsHMA3 gene sequence obtained in this study was completely consistent with CsHMA3a. The cDNA [GenBank: JX402100] was 2286 bp and the CsHMA3a encoded polypeptides of 762 amino acids in length and showed 96.5% identity with CsHMA3b [GenBank: KJ818901], while the presence of a stop codon at a position of 75 amino acids in length in CsHMA3c [GenBank: KJ818902] was observed (Additional file 2: Figure S2).
CsHMA3 belongs to the P1B-ATPases and includes the predicted eight transmembrane domains and the key identifying structures of P1B-type ATPases found in other subfamily members such as Arabidopsis and Thlaspi (Additional file 2: Figure S2), suggesting parallel functions in these species [17, 22, 26].
Expression pattern of CsHMAs
To investigate whether Camelina HMAs (heavy metal P1B-ATPases) can enhance functions in metal accumulation or tolerance, we first analyzed the expression patterns of eight HMA genes in Camelina (root, leaf, stem, flower, and silique) under normal conditions by reverse transcription (RT)-PCR. CsHMA primers were designed by multiple sequence alignment (Clustal W) in order to amplify a specific cDNA fragment of each gene. The PCR products were sequenced to confirm that only the transcriptions of the consistent gene were amplified. CsHMA3 and CsHMA7 genes were expressed in all organs. Expression of CsHMA1, CsHMA6, and CsHMA8 genes appeared to be weak only in siliques. CsHMA4 and CsHMA5 were mainly detected in the root, stem, and flower. The CsHMA2 expression pattern was similar to those of CsHMA4 and CsHMA5 except for low transcript level in the root (Figure 3A). In Arabidopsis, HMA1 to HMA4, which transport Zn2+, Cd2+, Pb2+, and Co2+, and AtHMA5 to AtHMA8, which transport Cu+ and Ag+ were identified by several studies [15–19]. Therefore, we mainly analyzed the expression profiles of CsHMA1, 2, 3, and 4 as potential candidate genes for the specific transport or storage of Cd, Pb, Zn, and Co. RT-PCR analyses were performed using RNAs isolated from the roots and leaves of Camelina grown in hydroponic culture containing either 50 μM Cd, 500 μM Pb, 500 μM Zn, or 100 μM Co. In some cases up-regulated expression levels of a particular gene could be found. The expression of CsHMA1 and CsHMA2 showed elevated levels in Pb-treated roots, and CsHMA3 seemed to be induced in roots and leaves from 6 hours after Pb treatment. Although the CsHMA4 genes exhibited unaltered expression levels in roots during Pb treatment, an induction in the leaves was shown from 6 hours after Pb treatment (Figure 3B). Among CsHMA1 to CsHMA4, the expression levels of CsHMA1 and CsHMA3 were more highly regulated by Pb, Zn, Co, and Cd (Figure 3B) than others. This data raised the possibility that CsHMA1 and CsHMA3 can be used as potential candidates for phytoremediation in future studies. Since HMA3 has been identified in various plants and is an important key factor that enhances metal uptake and effectively detoxifies toxic metals by compartmentation into vacuoles [22, 27], we focused on the CsHMA3 gene in this study.
Expression profile of CsHMAs. RT-PCR analyses were performed using RNAs isolated from various tissues in normal condition (A) or from roots and leaves 0, 6, 12, 24, and 48 hours after μM Cd, 350 μM Pb, 350 μM Zn, and 50 μM Co treatment (B). Plants grown hydroponically were used in these experiments. Root (Ro), leaf (Le), stem (St), flower (Fl), silique (Si).
Functional characterization of CsHMA3 by heterologous assay in yeast
To investigate whether CsHMA3 could function in metal tolerance, we performed a yeast functional complementation. The empty vector pYES and the pYES vector containing CsHMA3 were used to transform the yeast Δycf1 (Cd/Pb-sensitive mutant) or Δzrc1 (Zn-sensitive mutant). The phenotype of Δycf1 is caused by the loss of a yeast cadmium factor 1 (YCF1), a vacuolar glutathione S-conjugate of the ATP-binding cassette (ABC) transporter family associated with multidrug resistance in Saccharomyces cerevisiae. YCF1 has been reported to contribute to Cd and Pb resistance [28, 29]. The zinc resistance conferring1 (ZRC 1) in S. cerevisiae, a member of the cation diffusion facilitator (CDF) family that is involved in zinc efflux and compartmentalization, is known to contribute to zinc tolerance. In addition, Δzrc1 strains are hypersensitive to zinc [30]. Our results confirm that these mutants have a higher metal-sensitive phenotype compared with the wild-type yeast strain (Figure 4A). Although CsHMA3 expression in the wild-type strain could not enhance the tolerance of Cd, Pb, and Zn, the Δycf1 or Δzrc1 mutant strain yeast expressing CsHMA3 was able to complement the growth defect in the presence of 10 μM Pb and 5 mM Zn, but not in the Cd medium (Figure 4B). Rather the expression of CsHMA3 in the wild-type strain and Δycf1 increases sensitivity to Cd. According to a previous report [31], the expression of AtHMA3 in metal-sensitive yeast strains slightly increases tolerance to Cd and Pb, but not to Zn. The phenotype of CsHMA3-expressing strains was partially consistent with the expectation according to AtHMA3 function. Although it is difficult to determine precise metal substrates for CsHMA3 at the current stage, our result suggested that the expression of CsHMA3 is indirectly responsible for the increased Pb and Zn tolerance in metal-sensitive yeast strains.
Functional assay of CsHMA3 of the wild type and mutants in yeast. Confirmation of Cd and Pb sensitivity of Δycf1, and Zn sensitivity of Δzrc1 mutant strain compared with those of wild-type (BY4741) yeast strain (A). Cell growth of the Δycf1 or Δzrc1 mutant strain was transformed with empty vector pYES2, CsHMA3 (B). Yeast cells grown in the log phase were diluted to an OD600 of 1, 10-1, 10-2, 10-3, and 10-4, and 5 μL of each dilution were spotted onto the 1/2 YPD (A) or LPM medium containing galactose (B) without or with added CdSO4, PbNO3, and ZnSO4. The plates were incubated at 30°C for four days before photography. The yeast cells were grown in the presence of 25 μM Cd, 10 μM Pb, and 5 mM Zn. The photographs shown are representative results of independent experiments that were performed three times with reproducible results.
Overexpression of CsHMA3 in Camelinaimproved Cd, Pb, and Zn tolerance
To test whether CsHMA3 can be used to develop plants with enhanced metal tolerance, we generated transgenic Camelina expressing a 35S promoter:: CsHMA3, using a gateway cloning system. After confirmation of CsHMA3 transcript levels of five CsHMA3a independent transgenic T2 plants (H3-1, 3-2, 3-3, 3-5, and 3-7) and the wild type determined by RT-PCR using a 35S promoter primer and CsHMA3 specific primer (Additional file 3: Figure S3A), we selected the H3-1 and H3-3 CsHMA3 transgenic lines, which showed a much higher transcript level as compared to other lines, and then renamed them OX1 and OX2, respectively. In addition, OX3 is H3-2 and OX4 is H3-7. Differences in the phenotypes of wild-type and CsHMA3-overexpressing plant leaves were observed. Transgenic lines have more rounded-type leaves, whereas wild-type leaves are sharp (Additional file 3: Figure S3B).
To evaluate heavy metal tolerance of CsHMA3 transgenic lines, seeds of wild-type and two transgenic lines were germinated and grown on agar plates containing 25, 50, or 100 μM Cd, 250, 350, or 500 μM Pb or Zn, and 50 or 100 μM Co for one week. In the case of Pb treatment, we used modified medium without phosphate for control and Pb treatment to avoid Pb precipitation. No differences occurred in the germinations between the wild-type and transgenic lines in treatments compared to the control. The root growth and shoot fresh weight were measured one week after treatment. The transgenic lines grew better than the wild-type lines under Cd, Pb, and Zn stress (Figure 5). In particular, the transgenic lines showed enhanced Pb tolerance in a wide range of Pb concentrations (Figure 5). However, in the control medium and Co-treated medium, the growth of each plant was similar (Additional file 4: Figure S4). In addition, the contribution of overexpression of CsHMA3 to the enhanced Pb tolerance of Camelina was further evaluated by measuring the electrolyte leakage from the leaves of the plants (Figure 6). The leaves from the CsHMA3 transgenic line showed a lower rate of electrolyte leakage than that observed in the leaves from the wild types.
Effect of metal treatments on root growth and shoot fresh weight in transgenic lines. Photographs of Camelina wild-type and two CsHMA3-overexpressing lines grown on agar plates containing different concentrations of Cd, Pb, and Zn stress for one week (A). Root growth rate (B) and fresh weight (C) of the plants were measured at the seventh day after heavy metal treatment. For the Pb treatment, we used control medium without phosphate. Data represent means ± SD (n = 30). Asterisks in the figure indicate significant difference from wild type at *P < 0.05, **P < 0.01, and ***P < 0.001 by Dunnett’s test.
Effect of metal treatments on the electrolyte leakage of leaves in transgenic lines. Electrolyte leakage was expressed in percentage (%) of total electrolyte leakage (A). Data represent means with ± SD (n = 3). Different lowercase letters in the figure indicate significant difference at P < 0.05 by Tukey’s test. Two-week-old plants were hydroponically grown and treated with 50 μM Cd, 350 μM Pb, 350 μM Zn, and 50 μM Co for seven days (B). For Pb treatment, plants were transferred to phosphate-free nutrient solution at least 12 hours prior to treatment.
CsHMA3 overexpressing transgenic Camelinahave increased Pb and Zn contents
To analyze the heavy metal uptake ability of the CsHMA3 transgenic line, we measured the Cd, Pb, Zn, and Co contents in wild-type and overexpressing lines treated with 50 μM Cd, 350 μM Pb, 350 μM Zn, and 50 μM Co in hydroponic culture for one week. Based on the analysis of heavy metal tolerance between the wild-type and CsHMA3 transgenic lines, we determined appropriate concentrations of heavy metal treatments. The results showed that Pb and Zn contents in shoots of the transgenic line were higher than those of the wild type, but no significant differences were observed in the Cd contents of wild-type and transgenic plants, and the Co content in the shoots of the transgenic lines was lower than that of the wild type (Figure 7). In the roots, no difference in Pb and Zn contents between the transgenic and wild-type lines was observed. However, in the case of Cd and Co treatments, the Cd and Co contents of OX2 were higher than those of the wild type and OX1. The metal translocation ratios were calculated from these metal contents data. The Pb and Zn translocation ratios in the two transgenic lines were higher than those of the wild type. No significant difference was observed in the Cd translocation ratios between the transgenic and wild-type lines, but the Co translocation ratios of the transgenic lines were significantly lower compared to those of the wild type (Figure 7). These results suggest that overexpression of CsHMA3 affects the ability of Pb and Zn ion uptake and translocation in Camelina.
Metal contents in roots and shoots and the translocation ratio in the transgenic lines. Three-week-old plants were grown hydroponically and treated with 50 μM Cd, 350 μM Pb, 350 μM Zn, and 50 μM Co for one week. Data represent means ± SD (n = 3). Asterisks in the figure indicate significant difference from wild type at *P < 0.05 by Dunnett’s test.
Morphological analysis of CsHMA3overexpressing transgenic Camelina
As shown in Additional file 3: Figure S3B, the CsHMA3 transgenic lines exhibited leaves with a more rounded shape compared to the wild-type leaves. To test whether differences appeared in the leaf shape and phenotype between wild-type and CsHMA3 transgenic lines at the vegetative (Figure 8) and reproductive stages (Additional file 5: Figure S5), the length, width, angle, number of leaves, and stem length of plants were measured. In both growth stages, remarkable differences in phenotype between the wild-type and CsHMA3 transgenic lines were observed. Although no difference was observed in the length of leaves between all lines, the width of leaves in the transgenic lines was slightly greater than that of the wild type. In addition, the leaf angle of the CsHMA3 transgenic lines was markedly greater than that of the wild type. Each leaf angle was measured in the longitudinal and both lateral-end positions. In order to better characterize leaf shape, the leaf index was calculated by ratio of length to width in a leaf blade area. At the vegetative stage, the wild-type and CsHMA3 transgenic lines had 4.81 ± 0.16 and 3.68 ± 0.03 (mean ± SE of four transgenic lines) leaf index values, respectively. A value close to 1.0 is indicative of increased leaf roundness (Figure 8C). Indeed, the width size (in centimeters) of leaves in the CsHMA3 transgenic lines was 3.23 ± 0.03 and that of the wild type was 2.5 ± 0.13, but there was no significant difference in the leaf length between the wild-type and CsHMA3 transgenic lines (Figure 8B). This result indicates that the leaf shape of transgenic lines was rounder than that of the wild type. We also found a similar pattern of leaf index between the wild-type and CsHMA3 transgenic lines at the reproductive stage (Additional file 5: Figure S5). Furthermore, the leaf numbers of the CsHMA3 transgenic lines were greater than those of the wild type, but no difference in stem length between the wild-type and CsHMA3 transgenic lines was observed in both growth stages (Figure 8; Additional file 5: Figure S5).
Different phenotypes of leaves between wild-type and CsHMA3-overexpressing plants at the vegetative stage. Comparison of shape of leaves from four-week-old plants in hydroponic culture (A). Length of fifth leaf was measured in the leaf-length (longitudinal) and leaf-width (lateral) directions (B) and each leaf index was then determined by the ratio of length to width in a leaf blade (C). A value close to 1.0 is indicative of increased leaf roundness. Comparison of leaf angle (D). Each leaf angle was measured in the longitudinal and both lateral-end positions. Leaf count number (E). Leaves were collected from the same plants that were used for investigation of other leaf phenotypes. Measurement of stem length (F). Data represent means ± SD (n = 5). Different lowercase letters in the figure indicate significant difference at P < 0.05 by Tukey’s test.
Expression pattern of leaf shape related genes between wild-type and CsHMA3-overexpressing transgenic Camelina
In order to find some clue to the causes of changes in the leaf shape of CsHMA3-overexpressing transgenic Camelina, the need arose to test whether the expression of leaf shape related genes is affected by the overexpression of CsHMA3 in Camelina. To respond to this question, a targeted expression analysis of several genes that are involved in cell polarity was performed. Representatively, expression patterns of ANGUSTIFOLIA (AN) and ANGUSTIFOLIA3 (AN3), which are leaf-width direction regulators, and ROTUNDIFOLIA 3 (ROT3), ROTUNDIFOLIA 4 (ROT4), LONGIFOLIA1 (LNG1), and LONGIFOLIA2 (LNG2), which are leaf-length direction regulators, were investigated. The expression of a cell division regulator (CYCD3) was also checked as a means to evaluate whether the modification of cell division could be related to the altered leaf shape in CsHMA3 transgenic lines. Interestingly, transcript levels of cell division regulator and leaf-length direction regulators were induced in CsHMA3 transgenic lines at the vegetative stage (Figure 9). No significant differences were observed in the expression level of cell proliferation regulator genes between the wild-type and CsHMA3 transgenic lines. These results suggested that the induction of cell division regulator and leaf-length direction regulator transcripts was associated with alterations of leaf -width in CsHMA3 transgenic lines (Figure 9).
Gene expression of leaf shape related genes between wild-type and CsHMA3 -overexpressing transgenic Camelina.Total RNA was extracted from the fifth leaves in wild-type and CsHMA3 transgenic lines at the vegetative stage. Quantitative real-time PCR was performed to compare the expression levels of the CsCYCD3, CsAN, CsAN3, CsLNG1, CsLNG2, and CsROT3 genes. Expression (fold) was calculated by relative comparison to the expression of the individual genes in the wild type. CsActin was used as an internal control for normalization. Values are the averages of three replicates ± SE. Asterisks in the figure indicate significant difference from wild type at *P < 0.05, **P < 0.01, and ***P < 0.001 by Dunnett’s test.
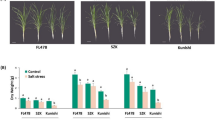
Analysis of productivity index of CsHMA3-overexpressing transgenic Camelinatreated with heavy metals in a greenhouse
In order to confirm whether soil-grown CsHMA3-overexpressing transgenic Camelina can also maintain the capacity of heavy metal tolerance for a longer period (90 days), we compared the productivity index between the wild-type and two transgenic lines. The productivity index includes the following: dry weight (g), height (cm), total number of siliques per plant, total seed weight (g), total seed number, and weight (g) per thousand seeds. When plants were grown under control conditions, no significant differences in the six productivity indexes were observed between the wild-type and the two transgenic lines. In the Cd, Pb, and Zn treatment, the dry weight of the two transgenic lines was higher by about 1.3- to 2-fold than that of the wild type, but not in the Co treatment. This is consistent with the heavy metal tolerance phenotype of transgenic lines observed from the vertical plate containing Cd, Pb, and Zn. In all the treatments, the total seed weight and total seed number of the two transgenic lines were significantly greater by about 1.5- to 3-fold compared to the wild type, but no significant difference was observed in weight per thousand seeds between the wild-type and the two transgenic lines (Table 1).
Discussion
Camelina is rich in omega-3 fatty acids, and various stages of research and development are currently being carried out on the use of Camelina as a biofuel. The production and utilization of Camelina as a biofuel have been limited because of the lack of definite knowledge about resistance to abiotic stresses, despite Camelina’s advantages of being able to adapt to a wide range of climates and being able to grow in barren environments that are usually unsuitable for food crops. Even though some research groups have reported that Camelina has resistance to diseases, pests, low temperature, and drought stress [6, 32–34], to the best of our knowledge, there has been no previous report on heavy metal stress tolerance.
It has been suggested that rapeseed, known as the representative oilseed crop, can possibly be used as a heavy metal extracting plant for phytoremediation [35–37]. To find out whether Camelina also has this possibility, we compared the metal tolerance and accumulation between Camelina and rapeseed. In a root elongation test under heavy metal stress, Camelina plants showed more Cd and Pb tolerance than rapeseed (Figure 1). When plants were exposed to heavy metal stress conditions, a negative effect of heavy metals on membrane integrity and cell permeability occurred and electrolytes were released to the outside of the membrane [38]. Measurement of electrolyte leakage has been used to quantify damages to cell membranes in several abiotic stresses such as drought, chilling and torrid conditions, air pollution, salt stress, acid conditions, and heavy metal exposure [39]. In an electrolyte leakage test, we treated both plants with Cd, Pb, Zn, and Co for five days. Camelina leaf showed significantly lower electrolyte leakage level than that of rapeseed leaf under Pb stress, but not under other metal stresses. Furthermore, the Pb, Zn, and Co contents in the Camelina shoot were higher than those of rapeseed (Figure 2). The results show that Camelina has similar or more tolerance depending on metal species and greater heavy metal translocation ability compared with rapeseed. Therefore, it is suggested that Camelina is more suitable than rapeseed to use for phytoremediation when considering heavy metal tolerance and accumulation.
Plants contain metal chelators and transporters that regulate metal homeostasis. The HMA3 gene is one of several potential candidates involved in mediating metal-hypertolerant and metal-hyperaccumulating traits, and encodes proteins belonging to the PIB-type. In addition, high HMA3 expression levels in shoots of a hyperaccumulator (T. caerulescens) have been reported. As a result, metal compartmentation into leaf vacuoles is enhanced, elevating metal translocation [22]. We determined the expression patterns for eight members of P1B-ATPases in Camelina (root, leaf, stem, flower, and silique) under normal conditions. Among them, the CsHMA3 gene was expressed in all organs. CsHMA2 and CsHMA4, known as a physiological master switch in metal hyperaccumulation, were detected in the stem and flower (Figure 3A). In addition, CsHMA3 showed remarkably elevated levels in Pb-treated leaves (Figure 3). We have hypothesized that the high expression of the CsHMA3 gene might improve heavy metal tolerance and accumulation, especially for Pb. Therefore, we attempted to determine how the CsHMA3 protein can change the ability of heavy metal uptake and translocation in plants. Also, the expression levels of HMA1 were highly regulated by Pb, Zn, Co, and Cd (Figure 3B). These data also raised the possibility that CsHMA1 can be used as a potential candidate gene for phyoremediation in future studies.
In this study, we show that CsHMA3-overexpressing Camelina plants show tolerance against Cd, Pb, and Zn, but not against Co, and that they enhance the accumulation of Pb and Zn in Camelina shoots (Figures 5, 6, and 7). We provide several lines of evidence to implicate CsHMA3 in heavy metal tolerance in Camelina. First, the Δycf1 or Δzrc1 mutant strain yeasts expressing CsHMA3 were able to complement the growth defect in the presence of 10 μM Pb and 5 mM Zn, but not in the Cd medium (Figure 4). The pattern of growth of transformed yeast cells showed results partly similar to those of the AtHMA3-transgenic yeast cell [31]. Functional expression of AtHMA3 phenotypically complements the Cd/Pb-hypersensitive yeast strain Δycf1, but not the Zn-hypersensitive mutant Δzrc1 [31]. Although the complementation of yeast growth defect to Cd failed in the expression of CsHMA3, our result indicated that CsHMA3 complemented the Pb and Zn sensitivity of yeast mutants. To further support this hypothesis, an additional experiment aiming at the determination of the metal concentration in the yeasts experiments was required. It may be possible to explain that the opposite phenotype of CsHMA3-expressing yeast under Cd stress and Pb/Zn stresses was caused by either internal sequestration or metal efflux out of the cell dependent on the metal species. Second, we demonstrate that CsHMA3-overexpressing lines grow better than wild-type lines in Cd-, Pb-, or Zn-containing medium or solution (Figure 5A and Figure 6B), and the experimental results can be explained well by differences between root length, fresh weight, and electrolyte leakage between CsHMA3 transgenic lines and the wild type (Figures 5B, C, and 6A). Third, as shown in Figure 7, it is worth noting that CsHMA3 transgenic lines mediate efficient translocation of the absorbed Pb and Zn from root to shoot. Although we are not certain about the mechanism of increased translocation in the CsHMA3 transgenic lines, we propose that CsHMA3 is likely to act as a short-term storage “filter” in the root, unlike AtHMA3 [40, 41], thereby less limiting heavy metal translocation to the shoot. This is similar to the proposal for the function of TcHMA3 in T. caerulescens [22].
However, CsHMA3 and TcHMA3 seem to have different metal substrate specificities. Overexpression of CsHMA3 increased the Pb and Zn tolerance and accumulation, but not those of Co. Enhanced tolerance of Cd was also observed in transgenic plants, but no difference in Cd accumulation between wild-type and transgenic lines was observed (Figures 4, 5, 6 and 7). When TcHMA3 was expressed in either yeast or Arabidopsis, it only enhanced tolerance of Cd, but not of Pb and Co. Furthermore, in genome-wide association studies, it was determined that HMA3 drives natural variation occurring in Cd concentration of leaf in A. thaliana, and grafting indicated that HMA3 functions in the root to govern shoot Cd translocation [42]. This is consistent with the OsHMA3 limiting translocation of Cd through the xylem into the shoot [43, 44]. These results suggest that other HMA homologs in various species have different functions with respect to substrate specificity and toxic metal sequestration. It has been proposed that the metal specificity of P1B-ATPases is indicated by conserved amino acids located in transmembrane domain (TMD) 6 at the CPx or SPC sequences, and by signature sequences in TMD7 and TMD8 in P1B-ATPases [45]. By analyzing all available P1B-ATPase protein sequences, and searching for similarities and conserved amino acid sequences in TMD6, TMD7, and TMD8, signature sequences were identified and the P1B-ATPases were divided into four subgroups with distinct metal specificity (1B-1, Cu+/Ag+; 1B-2, Zn2+/Cd2+/Pb2+/Co2+; 1B-3, Cu2+/Cu2+/Ag2+; 1B-4, Cu2+) and two subgroups (1B-5 and 1B-6) [17, 45]. Although these three specific amino acid regions are identical in HMA3 proteins of Camelina, T. caerulescens, A. thaliana (Ws), and A. halleri, the metal-substrate specificity of these plants is not completely matched [22]. Thus, the specific factor related to determining metal-substrate specificity, based on additional structural considerations in the N- and/or C-terminal ends of CsHMA3, requires further study.
Here, we also found that CsHMA3-overexpressing Camelina plants clearly exhibited a wider and more orbicular leaf blade than the wild type, and leaf numbers in the CsHMA3 transgenic lines were also greater than in the wild-type lines at both vegetative and reproductive growth stages (Figure 8). This indicated that these leaf phenotypes of CsHMA3 transgenic lines did not correlate to any difference in growth stages. The shapes and sizes of leaves are crucial factors influencing the life of plants, because leaves are photosynthetic organs. To absorb enough light energy, leaves need to be as wide as possible. However, the leaf area is principally regulated by the availability of water [46]. The formation of leaf shape and size is affected by cell differentiation on the leaf surfaces during the stages of leaf morphogenesis and lateral, two-dimensional expansion of the leaf blade [47]. Generally, these steps are regulated by several leaf shape related genes [47, 48]. Arabidopsis leaf blades expand in two dimensions, which are described as leaf-length (longitudinal) and leaf-width (lateral) axes. The ratio of the two axes has been used as the standard to establish leaf blade shape [49]. The leaf-width (lateral) and leaf-length (longitudinal) axes of leaf expansion are regulated by the AN, and the ROT genes, respectively. In addition, AN3/GRF-INTERACTING FACTOR1 (AtGIF1) and SPIKE1 have been known as regulators in the leaf-width direction [50–53]. With respect to the regulation of leaf length, ROT3, ROT4, LNG1, and LNG2 are involved in the regulation of leaf length polarity growth [54–56]. Arabidopsis cold shock domain protein 3 (AtCSP3) may also function to regulate leaf-length direction [49]. The real-time (RT)-PCR data in Figure 9 suggest that a wider leaf shape observed in CsHMA3 transgenic lines might be caused by inductions in the transcript level of leaf-width direction regulator genes.
The ectopic expression of CsHMA3 all over the plant body likely modifies the concentration of an essential metal such as Zn in cellular compartments. This modification interacts with the whole plant’s metal-homeostasis regulation and with numerous pathways, leading to changes in the regulation of leaf development and metal translocation to shoots of CsHMA3 transgenic lines. Thus, we could not assert that these changes directly indicate the physiological function of the CsHMA3. To understand the exact role of CsHMA3, further experiments such as phenotypic analysis of a CsHMA3 knockdown plant and identification of its tissue and cellular location are required.
The aim of our study was to show if Camelina could be used for phytoremediation of metal-contaminated soil and in parallel for oil production. However, some crucial problems about the content of heavy metals in the oil obtained from Camelina plants grown in heavy metal contaminated areas remained unsolved. Further research to investigate if biofuels containing heavy metals exhaust poisonous gas is required. Also, it is crucial to determine whether overexpression of HMA3 affects the fatty acids composition, which is responsible for the fuel properties of biodiesel, in plants grown both under control conditions and in the presence of metals. The productivity index analysis of CsHMA3 transgenic and wild-type lines indicated that CsHMA3-overexpressing transgenic plants showed less inhibition of the production of silique by Cd and Pb stress; thus, the total seed number and total seed weight were higher than those of the wild type (Table 1). Even though no difference in total number of siliques was observed between the wild-type and the two transgenic lines under Zn and Co stress, the total seed number and total seed weight in the two transgenic lines were higher than those of the wild type, because there were significantly more seeds per silique. These observations indicate that the CsHMA3 gene can contribute to increasing the productivity of seeds in the heavy metal contaminated soil.
Conclusions
In conclusion, our data demonstrated that CsHMA3 affects the Cd, Pb, and Zn tolerance and the ability of heavy metal uptake and root-to-shoot translocation, especially for Pb and Zn in Camelina. This provides the possibility for phytoremediation of multiple metal contaminated sites. Also, overexpression of CsHMA3 resulted in larger leaf size compared with the wild type due to an induction of leaf shape related genes and showed an increasing productivity of seeds in the soil treated with heavy metal. This information will also be useful to improve productivity of bioenergy crops. Moreover, the heterologous overexpression of HMA3 and HMA4 in plants is probably a useful strategy to engineer altered metal distribution in tissues for biofortification or phytoremediation purposes [57].
Materials and methods
Plant material and growth conditions
Seeds of Camelina sativa L. cv. “CAME” and B. napus L. cv. “Youngsan” were grown at 22 ± 1°C under long days (16-/8-h day/night light with 200 μmol m-2 s-1 cycle). To analyze the germination rate and seedling growth under heavy metal stresses, plants were grown vertically on 125 × 125 × 20 mm polystyrene petri dishes containing half-strength Murashige and Skoog medium (Duchefa Biochemie; basal salts mixture) or modified nutrient medium (without phosphate to avoid Pb precipitation) containing 1.25 mM KNO3, 1.5 mM Ca(NO3)2, 0.75 mM MgSO4, 75 μM FeEDTA, 50 μM H3BO3, 10 μM MnCl, 2 μM ZnSO4, 1.5 μM CuSO4, and 0.075 μM (NH4)6Mo7O24 containing 0.8% (w/v) phytoagar (Duchefa Biochemie) and 1.5% (w/v) sucrose after sterilization with 75% ethanol for 10 min. CdCl2, Pb(NO3)2, ZnSO4, and CoCl2 were supplemented into the corresponding media at the concentrations indicated in the Figures 1, 5 and Additional file 4: Figure S4. To reduce the variation due to the precipitation of CdCl2, Pb(NO3)2, ZnSO4, and CoCl2, the plants were grown in the same plate and their growth was monitored for three weeks. A seed was regarded as germinated when the radicle protruded from the seed coat. Germination and growth assays were carried out on three replicates.
Measurement of electrolyte leakage
For the measurement of electrolyte leakage, leaves were excised and washed three times with distilled water. After removing the water with filter paper, the leaves were cut into small pieces (about 5 mm) and placed in flasks with 60 mL distilled water. The flasks were shaken at 26˚C for 2 hours [58], and the electrical conductivity of the solution (EC1) was measured using an IQ170 electrical conductivity meter (IQ Scientific Instruments, San Diego, CA, USA). Autoclaved samples (EC2) were measured to determine the maximum percentage of electrolyte leakage. The relative electrical conductivity (REC) was calculated as REC (%) = (EC1/EC2) × 100.
Expression analysis of CsHMAsand leaf shape related genes
To determine the expression of CsHMAs and leaf shape related genes by reverse transcription (RT)-PCR, the total RNA was obtained from tissues (flower, stem, leaf, and root) of four- or six-week-old Camelina grown on rockwool using the RNAiso Plus Kit (Takara code: D9108A, Shiga, Japan) and the RNeasy Plant Mini Kit (Qiagen, Valenica, CA) according to manufacturer instructions. The quantity of RNA was measured using a spectrophotometer (Nanovue, Daejeon, Korea), and its quality was checked using agarose gel electrophoresis. RNA was treated with RNase-free DNase-I (Ambion, Austin, TX) for 45 min followed by DNase-I removal as specified by the manufacturer. Two micrograms of DNA-free RNA were then reverse transcribed using the First-Strand Synthesis System (Invitrogen) for RT-PCR according to the manufacturer’s specifications. cDNA concentrations were normalized with CsActin or beta-tubulin. To amplify CsHMA1 to about eight cDNAs, PCRs were performed in a final volume of 20 μL using cDNA as templates in a PTC-100 Peltier Thermal Cycler (MJ Research, Waltham, MA) using specific primers (Additional file 6: Table S1). Cycling conditions were 5 min at 95°C, followed by 25 or 30 cycles of 15 s at 95°C, 30 s at 55.3 or 57.3°C, and 30 s at 72°C. At the end of the cycling, the samples were incubated at 4°C. For comparative quantification of leaf shape related genes, real-time quantitative PCR (qRT-PCR) was performed in a Bio-Rad CFX96 Real-Time PCR system (Bio-Rad Laboratories Inc., Hercules, CA, USA). The 20-μL PCR reaction mixture contained 4 μL of diluted cDNA, 10 μL of SYBR Premix (Takara), 2 μL of a mix of the primer pair, and 4 μL of water. The real-time PCR condition was followed by a three-step protocol with melting curve. The data were analyzed using the Bio-Rad CFX manager (version 1.0) after normalization to CsActin. Triplicate PCR and three biological replicates were analyzed. The primers used for this experiment are listed in Additional file 6: Table S1.
Vector construction and Camelinatransformation
☐Total RNA was isolated from two-week-old seedlings using the RNAiso Plus Kit (Takara code: D9108A, Shiga, Japan). RNA was treated with RNase-free DNase-I (Ambion, Austin, TX) for 45 min followed by DNase-I removal as specified by the manufacturer, and the cDNA was synthesized using the SMARTerTM RACE cDNA Amplication Kit (Clontech, Mountain View, CA). The full-length coding sequence of CsHMA3 was obtained using 3’ and 5’ rapid amplification of cDNA ends (RACE Technique, Clontech) according to the protocol of the SMART™ RACE kit (Clontech). Full-length cDNA was cloned using 5′-AAAAAGCAGGCTATGGTGGAAGGTGAAGAGACAAAG-3′ and 5′-AGAAAGCTGGGTTCACTTTTGTTGATCCTCCTTAGGGCC-3′ primers. Two recombination sites, attB1 and attB2 at the 5’-ends, were added to the forward and reverse primer, respectively. The pair of PCR primers was not specific for only CsHMA3a full sequence amplification. The PCR products were purified and cloned into the pGEM-T Vector System (Promega, Madison, WI). Seven clones were obtained and sequenced. Among them, sequences of four clones were consistent with each other. This sequence has been deposited in GenBank [GenBank:JX402100] (CsHMA3a). Subsequently, this gene was cloned in pDONR207 using the Gateway Cloning System (Invitrogen). The binary vector pCB302-3 [59] contained the promoter and terminator of the CaMV (35 s promoter) to overexpress a CsHMA3 cDNA in Camelina and was modified to be compatible with the Gateway system as described by the manufacturer. Clones from which the expected sequence was confirmed were transferred to the pCB302-3 vectors. The transformation of Camelina was conducted via vacuum infiltration while immersing flowers for 5 min in the Agrobacterium-containing solutions [60]. Camelina plants at the early flowering stages were used for transformation. To get a high ratio of transformation, plants were used three times for transformation at four-day intervals. Selection was performed in E. coli and A. tumefaciens strains with 60 μg/mL kanamycin and in plants with 50 mg/mL DL-phosphinotricin (PPT) (Duchefa, Harlem, The Netherlands). The frequency of transformation was about 0.5%.
Yeast transformation and Zn/Cd tolerance analysis
The wild-type S. cerevisiae strain BY4741 (EUROSCARF accession number Y0000; MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0), a BY4741-derived Cd/Pb-sensitive strain with Δycf1 (Y04069), and a Zn-sensitive strain with Δzrc1 (Y00829) were used for complementation analysis of CsHMA3. The pYES2-CsHMA3 expression constructs were generated by inserting CsHMA3 expression cassettes into the BamHI/XhoI sites of pYES2 (driven by the GAL1 promoter). The construct was then introduced into the wild-type yeast strain BY4741, the Cd/Pb-sensitive Δycf1 strain, and the Zn-sensitive Δzrc1 yeast strain using the lithium acetate method [61]. Yeast cells transformed with empty pYES2 were used as a control. Transformants were selected on 2% glucose, 0.67% Yeast Nitrogen Base (without amino acids, Difco), 0.16% Yeast Synthetic Drop-Out Medium Supplement Without Uracil (Sigma-Aldrich), and 1.5% bacto agar. For the metal tolerance assays, yeast cells grown in the log phase were diluted to an OD600 of 1, 10-1, 10-2, 10-3, and 10-4, and 5 μL of each dilution were spotted onto LPM medium containing galactose without or with added CdSO4, PbNO3, and ZnSO4. The plates were incubated at 30°C for four days before photography. The yeast cells were grown in the presence of 25 μM Cd, 10 μM Pb, and 5 mM Zn.
Measurements of metal contents
Plants were grown on rockwool in a 4-L container volume (six or nine plants per one container) containing aerated nutrient solution for about three to four weeks. The nutrient solution contained 1.25 mM KNO3, 1.5 mM Ca(NO3)2, 0.75 mM MgSO4, 0.5 mM KH2PO4, 75 μM FeEDTA, 50 μM H3BO3, 10 μM MnCl, 2 μM ZnSO4, 1.5 μM CuSO4, and 0.075 μM (NH4)6Mo7O24. The solutions were supplemented every day and changed once a week or to avoid the excessive depletion of any particular ion. For heavy metal treatments, plants were transferred to a solution with concentrations of CdCl2, Pb(NO3)2, ZnSO4, or CoCl2, as indicated in the Figures 2 and 7. In the case of Pb treatment, plants were transferred to phosphate-free nutrient solution at least 12 hours prior to treatment. After growing for seven days, plant tissues (root and shoot) were harvested, rinsed in ice-cold 1 mM tartarate solution, and blot-dried at 60°C for three days. Each dried sample was weighed and digested with 11 N HNO3 at 200°C using a microwave digestion system (MDS Ethos1, Milestone, Italy). Digested samples were diluted with 0.5 N HNO3 and then analyzed using inductively coupled plasma mass spectroscopy with an IRIS-AP apparatus (Thermo Scientific, Pittsburgh, PA). The rate of metal translocation was estimated as the percentage of metal content in the shoot compared with that in the whole plant [44].
Statistical analysis
Statistical analyses for biological data were carried out using the SPSS statistical software (SPSS, Chicago, IL, USA). Statistical differences between measurements (n = 3, 5, or 30) for different results were analyzed following Student’s t-test, Dunnett’s test, and Tukey’s test. Values of P < 0.05, P < 0.01, or P < 0.001 were considered statistically significant.
Abbreviations
- HMA:
-
heavy metal ATPase
- RACE:
-
rapid amplification of cDNA ends
- OX:
-
overexpression.
References
Sheehan J, Camobreco V, Duffield J, Graboski M, Shapouri H: Life Cycle Inventory of Biodiesel and Petroleum Diesel for Use in an Urban Bus. Final Report. 1998, National Renewable Energy Laboratory, Golden, Colorado, USA; NREL/SR-580–24089
Demirbas A: Global renewable energy projections. Energ Source Part B. 2009, 4: 212-224.
Saxena RC, Adhikari DK, Goyal HB: Biomass-based energy fuel through biochemical routes: a review. Renew Sust Energ Rev. 2009, 13: 167-178.
Banerjee R, Mishra G, Nigam S: Non-edible oil-seeds potential raw material for industry. J Sci Ind Res. 1983, 42: 686-
Knorzer K: Evolution and spreading of gold of pleasure (Camelina sativa S.L.). Ber Deutsch Bot Ges. 1978, 91: 187-195.
Putnam D, Budin J, Field L, Breene W: Camelina: a promising low-input oilseed. New Crops. 1993, New York: Wiley, 314-322.
Vollmann J, Damboeck A, Eckl A, Schrems H, Ruckenbauer P: Improvement of Camelina sativa, an underexploited oilseed. Progress in New Crops. 1996, Alexandria, VA: ASHS Press, 1: 357-362.
Zubr J: Oil-seed crop: Camelina sativa. Ind Crops Prod. 1997, 6: 113-119.
Zubr J, Matthaus B: Effects of growth conditions on fatty acids and tocopherols in Camelina sativa oil. Ind Crops Prod. 2002, 15: 155-162.
Pilgeram A, Sands D, Boss D, Dale N, Wichman D, Lamb P, Lu C, Barrows R, Kirkpatrick M, Thompson B: Camelina sativa, a Montana omega-3 and fuel crop. Issues in New Crops and New Uses. Edited by: Janick J, Whipey A. 2007, Alexandria, VA: ASHA Press, 129-131.
Pence NS, Larsen PB, Ebbs SD, Letham DL, Lasat MM, Garvin DF, Eide D, Kochian LV: The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci U S A. 2000, 97: 4956-4960.
Vassilev A, Schwitzguebel JP, Thewys T, van der Lelie D, Vangronsveld J: The use of plants for remediation of metal-contaminated soils. Scientific World J. 2004, 4: 9-34.
Reeves RD, Baker AJ: Metal-accumulating plants. Phytoremediation of Toxic Metals: Using Plants to Clean up the Environment. 2000, New York: Wiley, 193-229.
McGrath SP, Zhao F-J: Phytoextraction of metals and metalloids from contaminated soils. Curr Opin Biotechnol. 2003, 14: 277-282.
Rensing C, Sun Y, Mitra B, Rosen BP: Pb(II)-translocating P-type ATPases. J Bio Chem. 1998, 273: 32614-32617.
Eren E, Arguello JM: Arabidopsis HMA2, a divalent heavy metal-transporting PIB-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol. 2004, 136: 3712-3723.
Williams LE, Mills RF: P1B-ATPases - an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005, 10: 491-502.
Baekgaard L, Mikkelsen MD, Sorensen DM, Hegelund JN, Persson DP, Mills RF, Yang Z, Husted S, Andersen JP, Buch-Pedersen MJ, Schjoerring JK, Williams LE, Palmgren MG: A combined zinc/cadmium sensor and zinc/cadmium export regulator in a heavy metal pump. J Biol Chem. 2010, 285: 31243-31252.
Migeon A, Blaudez D, Wilkins O, Montanini B, Campbell M, Richaud P, Thomine S, Chalot M: Genome-wide analysis of plant metal transporters, with an emphasis on poplar. Cell Mol Life Sci. 2010, 67: 3763-3784.
Van Hoof NALM, Hassinen VH, Hakvoort HWJ, Ballintijn KF, Schat H, Verkleij JAC, Ernst WHO, Karenlampi SO, Tervahauta AI: Enhanced copper tolerance in Silene vulgaris (Moench) Garcke populations from copper mines is associated with increased transcript levels of a 2b-type metallothionein gene. Plant Physiol. 2001, 126: 1519-1526.
Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Kramer U: Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature. 2008, 453: 391-395.
Ueno D, Milner MJ, Yamaji N, Yokosho K, Koyama E, Clemencia Zambrano M, Kaskie M, Ebbs S, Kochian LV, Ma JF: Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 2011, 66: 852-862.
Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, Clarke C, Higgins EE, Huebert T, Sharpe AG, Parkin IAP: The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun. 2014, 5: 3706-
Rossi G, Figliolia A, Socciarelli S, Pennelli B: Capability of Brassica napus to accumulate cadmium, zinc and copper from soil. Acta Biotechnol. 2002, 22: 133-140.
Grispen VM, Nelissen HJ, Verkleij JA: Phytoextraction with Brassica napus L.: a tool for sustainable management of heavy metal contaminated soils. Environ Pollut. 2006, 144: 77-83.
Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB: Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003, 132: 618-628.
Park W, Ahn SJ: How do heavy metal ATPases contribute to hyperaccumulation?. J Plant Nutr Soil Sc. 2014, 177: 121-127.
Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA: A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci U S A. 1997, 94: 42-47.
Song WY, Sohn EJ, Martinoia E, Lee YJ, Yang YY, Jasinski M, Forestier C, Hwang I, Lee Y: Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat Biotechnol. 2003, 21: 914-919.
Kamizono A, Nishizawa M, Teranishi Y, Murata K, Kimura A: Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1989, 219: 161-167.
Gravot A, Lieutaud A, Verret F, Auroy P, Vavasseur A, Richaud P: AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett. 2004, 561: 22-28.
Hansen L: Intertribal somatic hybridization between rapid cycling Brassica oleracea L. and Camelina sativa (L.) Crantz. Euphytica. 1998, 104: 173-179.
Kim HS, Oh JM, Luan S, Carlson JE, Ahn SJ: Cold stress causes rapid but differential changes in properties of plasma membrane H+-ATPase of Camelina and rapeseed. J Plant Physiol. 2013, 170: 828-837.
Jang HY, Yang SW, Carlson JE, Ku YG, Ahn SJ: Two aquaporins of Jatropha are regulated differentially during drought stress and subsequent recovery. J Plant Physiol. 2013, 170: 1028-1038.
Marchiol L, Assolari S, Sacco P, Zerbi G: Phytoextraction of heavy metals by canola (Brassica napus) and radish (Raphanus sativus) grown on multicontaminated soil. Environ Pollut. 2004, 132: 21-27.
Nouairi I, Ammar WB, Youssef NB, Daoud DBM, Ghorbal MH, Zarrouk M: Comparative study of cadmium effects on membrane lipid composition of Brassica juncea and Brassica napus leaves. Plant Sci. 2006, 170: 511-519.
Bi R, Schlaak M, Siefert E, Lord R, Connolly H: Influence of electrical fields (AC and DC) on phytoremediation of metal polluted soils with rapeseed (Brassica napus) and tobacco (Nicotiana tabacum). Chemosphere. 2011, 83: 318-326.
Zeid IM, Ghazi S, Nabawy D: Alleviation of Co and Cr toxic effects on alfalfa. Intl J Agron Plant Prod. 2013, 4: 984-993.
Bajji M, Kinet J-M, Lutts S: The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 2002, 36: 61-70.
Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P: AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009, 149: 894-904.
Park W, Han K-H, Ahn SJ: Differences in root-to-shoot Cd and Zn translocation and by HMA3 and 4 could influence chlorophyll and anthocyanin content in Arabidopsis Ws and Col-0 ecotypes under excess metals. Soil Sci Plant Nutr. 2012, 58: 334-348.
Chao DY, Silva A, Baxter I, Huang YS, Nordborg M, Danku J, Lahner B, Yakubova E, Salt DE: Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet. 2012, 8: e1002923-
Ueno D, Yamaji N, Kono I, Huang CF, Ando T, Yano M, Ma JF: Gene limiting cadmium accumulation in rice. Proc Natl Acad Sci U S A. 2010, 107: 16500-16505.
Miyadate H, Adachi S, Hiraizumi A, Tezuka K, Nakazawa N, Kawamoto T, Katou K, Kodama I, Sakurai K, Takahashi H, Satoh-Nagasawa N, Watanabe A, Fujimura T, Akagi H: OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011, 189: 190-199.
Argüello JM: Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J Membrane Biol. 2003, 195: 93-108.
Tsukaya H: Leaf shape: genetic controls and environmental factors. Int J Dev Biol. 2005, 49: 547-
Barkoulas M, Galinha C, Grigg SP, Tsiantis M: From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol. 2007, 10: 660-666.
Byrne ME: Networks in leaf development. Curr Opin Plant Biol. 2005, 8: 59-66.
Yang Y, Karlson D: Effects of mutations in the Arabidopsis cold shock domain protein 3 (AtCSP3) gene on leaf cell expansion. J Exp Bot. 2012, 63: 4861-4873.
Folkers U, Kirik V, Schobinger U, Falk S, Krishnakumar S, Pollock MA, Oppenheimer DG, Day I, Reddy AR, Jurgens G, Hulskamp M: The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 2002, 21: 1280-1288.
Kim G-T, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H: The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J. 2002, 21: 1267-1279.
Qiu JL, Jilk R, Marks MD, Szymanski DB: The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell. 2002, 14: 101-118.
Kim JH, Kende H: A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci U S A. 2004, 101: 13374-13379.
Tsuge T, Tsukaya H, Uchimiya H: Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development. 1996, 122: 1589-1600.
Kim GT, Tsukaya H, Uchimiya H: The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998, 12: 2381-2391.
Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H: CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 2005, 41: 710-721.
Siemianowski O, Mills RF, Williams LE, Antosiewicz DM: Expression of the P(1B)-type ATPase AtHMA4 in tobacco modifies Zn and Cd root to shoot partitioning and metal tolerance. Plant Biotechnol J. 2011, 9: 64-74.
Yu X, Peng YH, Zhang MH, Shao YJ, Su WA, Tang ZC: Water relations and an expression analysis of plasma membrane intrinsic proteins in sensitive and tolerant rice during chilling and recovery. Cell Res. 2006, 16: 599-608.
Xiang C, Han P, Lutziger I, Wang K, Oliver DJ: A mini binary vector series for plant transformation. Plant Mol Biol. 1999, 40: 711-717.
Bechtold N, Pelletier G: In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998, 82: 259-266.
Gietz D, St Jean A, Woods RA, Schiestl RH: Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20: 1425-
Acknowledgements
This study was supported by the World Class University project (R31-2009-000-20025-0) funded by the Ministry of Education, Science, and Technology, Korea. We would like to thank Prof. John E. Carlson (Pennsylvania State University) for providing wild-type Camelina seeds (accession PI 650140 “CAME”). We thank Dr. J.Y. Kim and Prof. K.H. Han (Michigan State University) for providing the binary vector pCB302-3. We also thank Dr. K.J. Kwak (Chonnam National University) for helping to generate the pYES-CsHMA3 construct.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WP planned the study, carried out the experiment, and drafted the manuscript. SJA planned the study and revised the manuscript. YF performed the investigation of productive parameters in the wild-type and CsHMA3 transgenic lines. All authors read and approved the final manuscript.
Electronic supplementary material
13068_2014_494_MOESM1_ESM.docx
Additional file 1: Figure S1: Effect of metal treatments on the electrolyte leakage of the leaves of Camelina and rapeseed. Four-week-old plants were grown in nutrient solution hydroponically and treated with 50 μM Cd, 500 μM Pb, 500 μM Zn, and 100 μM Co for five days. In the case of Pb treatment, plants were transferred to phosphate-free nutrient solution at least 12 hours prior to treatment. Data represent means ± SD (n = 6). Asterisks in the figure indicate significant difference between Camelina and rapeseed subjected to the same treatment at *P < 0.05 by Student’s t-test. (DOCX 30 KB)
13068_2014_494_MOESM2_ESM.docx
Additional file 2: Figure S2: Comparison of the amino acid sequences of three CsHMA3 copies according to Clustal W multiple alignment (version 1.83). The gray box shows metal binding domain (MBD), and lines over sequence indicate putative transmembrane domain (TMD), P-type ATPase signature (PAS), and HMA signature (HMAS). The red arrow indicates the position of the stop codon observed in CsHMA3c. (DOCX 59 KB)
13068_2014_494_MOESM3_ESM.docx
Additional file 3: Figure S3: Confirmation of CsHMA3 overexpression lines. CsHMA3 transcript levels of five CsHMA3 transgenic T2 plants (H3-1, 3-2, 3-3, 3-5, and 3-7) and wild type were determined by RT-PCR (A). Different phenotypes of leaves between wild-type and CsHMA3-overexpressing plants (B). (DOCX 452 KB)
13068_2014_494_MOESM4_ESM.docx
Additional file 4: Figure S4: Effect of Co stress on root growth in Camelina wild-type and transgenic lines. Seeds were germinated and grown on agar plates containing different concentrations of heavy metals for seven days. Photographs taken of Camelina wild-type and two CsHMA3-overexpressing lines grown on agar plates containing different concentrations of Co for one week (A). Root growth rate (B) of the plants was measured at the seventh day after heavy metal treatment. Data represent means ± SD (n = 30). (DOCX 933 KB)
13068_2014_494_MOESM5_ESM.docx
Additional file 5: Figure S5: Different phenotypes of leaves between wild-type and CsHMA3-overexpressing plants at the reproductive stage. Comparison of leaf shape of tenth leaf from plants 42 days after germination (DAG) in hydroponic culture (A). Leaf length was measured in the leaf-length (longitudinal) and leaf-width (lateral) directions. Comparison of leaf index (B). Each leaf index was determined by the ratio of length to width in a leaf blade. A value close to 1.0 is indicative of increased leaf roundness. Comparison of leaf angle (C). Each leaf angle was measured in the longitudinal and both lateralend positions. Leaf count number (D). Leaves were collected from the same plants that were used for investigation of other leaf phenotypes. Measurement of stem length (E) (n = 5). Different lowercase letters in the figure indicate significant difference at P < 0.05 by Tukey’s test. (DOCX 109 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Park, W., Feng, Y. & Ahn, SJ. Alteration of leaf shape, improved metal tolerance, and productivity of seed by overexpression of CsHMA3 in Camelina sativa. Biotechnol Biofuels 7, 96 (2014). https://doi.org/10.1186/1754-6834-7-96
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1754-6834-7-96