Abstract
Cotton is an international agricultural commodity and the main cash crop of Pakistan of which quality and quantity are subject to various whims of nature. Climate change, insect pest complex, and weeds are reducing its productivity. Here, we have developed triple gene cotton containing EPSPS gene along with two Bt toxin genes Cry1Ac and Cry2Ab using a strategy where all three genes are cloned in the same T-DNA, followed by successful cotton transformation via Agrobacterium-mediated transformation. This strategy has been developed to help cotton breeders in developing new cultivars by incorporating these genes into the non-transgenic or single Bt (Cry1Ac) gene cotton background where all three genes will inherit together. The expression of all three proteins was confirmed through immunostrips and was quantified through enzyme-linked immunosorbent assay (ELISA). The spatio-temporal expression of Bt protein in different parts of triple gene NIBGE cotton plants was determined. Maximum expression was found in leaves followed by seeds and boll rinds. Insect bioassays with cotton bollworms (Helicoverpa armigera), armyworms (Spodoptera litura), and pink bollworms (Pectinophora gossypiella) showed more than 90% mortality. The best performing line (NIBGE-E2) on the basis of spatiotemporal expression, glyphosate assays, and insect mortality data, was used for event characterization by using the genome sequencing approach. The event was successfully characterized and named NIBGE 20-01. A diagnostics test based on event-specific PCR was developed and its ability to distinguish NIBGE 20-01 event from other commercial transgenic cotton events was confirmed. To confirm stable expression of all three proteins in the field conditions, homozygous transgenic lines were grown in the field and the expression was confirmed through immunostrip assays. It was found that all three genes are expressed under field conditions. To show that all three genes are inherited together upon crossing with local elite cotton lines, the F1 generation was grown under glasshouse and field conditions. The expression of all three genes was confirmed under field conditions. Our results showed that transgenic cotton with three genes cloned in the same T-DNA can express all genes and can be conveniently transferred into elite cotton lines through a single cross.
Similar content being viewed by others
Introduction
Cotton (Gossypium hirsutum L.) being an essential fiber-producing crop globally is the main cash crop of Pakistan. It is cultivated on 2.6 million hectares and helps in engaging a substantial number of Pakistani populations towards economic activity. Pakistan is the fifth-largest producer of cotton after China, India, the USA, and Brazil. Cotton has a 0.8% share in the country’s GDP and contributes 4.1% in agricultural value addition1. Production of cotton is hampered by many biotic and abiotic factors. Biotic stresses mainly involve the attack of chewing insect pests like cotton bollworms (Helicoverpa armigera), spotted bollworms (Earias insulana), armyworms (Spodoptera litura), and pink bollworms (Pectinophora gossypiella)2 in addition to sucking insect pests including whiteflies (Bemisia tabaci)3, aphids and others4,5. Many abiotic factors like drought, salinity, high temperature, etc. also contribute to lowering yields. In Pakistan Bt cotton containing the Cry1Ac gene was formally approved for cultivation in 20106,7 which showed an increase in the production of cotton. The benefits of Bt cotton introduction among farmers in terms of minimizing pesticide use to control bollworms and decrease in the import bill of pesticides are well documented. Unfortunately, the success was not sustained due to the emergence of several insect pests which resulted in increased use of chemical pesticides. Recently it was found that pink bollworm has developed resistance against Bt cotton expressing the cry1Ac gene (Siddiqui et al., data not published) in Pakistan. The resistance breakdown was confirmed by field surveys and field trials. The mutations in the cadherin gene associated with resistance development in pink bollworms have been confirmed. Several known and novel mutations in the cadherin gene were detected in pink bollworms collected from across Pakistan. The extensive use of pesticides to control pink bollworms has not only increased the cost of cotton production but is also causing damage to the environment as well as flared-up whitefly population, probably because of the use of pesticides that promote whiteflies.
Current insect pest management strategies usually involve the use of chemicals such as pesticides which are associated with effects on human health, the environment8, and development of pesticide resistance among insect populations9,10,11. The benefits of GM crops are enormous and are one of the top most leading technologies in the world with exceptional adaptability among the farmers. GM crops adaptability among farmers is increasing every year which can be assessed by the fact that in 2005 the area under GM crops was just 90 million hectares12, which increased to 191 million hectares in 201813. Bt cotton was first introduced in 199614 and it played a vibrant role in enhancing the production of cotton but with time, production of cotton is threatened as the insects are developing resistance15,16,17 against the Bt cotton18 expressing Cry1Ac19,20 and Cry2Ab21,22,23. Up till now, a total of fifteen insect species are reported that developed resistance against the Bt toxins20 of which only five were reported until 201219. In Pakistan, most of the cultivated Bt cotton varieties had low Bt toxin expression levels and refuge plan was not properly followed which imposed high selection pressure on the chewing insect pests, leading to resistance development15,24,25,26,27,28. However, our trials data (data not published) from five majorly cotton growing areas of Punjab and Sindh province showed that double gene Bt cotton expressing Cry1Ac and Cry2Ab is still very much resistant and provides protection against all kinds of chewing insect pests including H. armigera, S. litura, and P. gossypiella.
Weeds are also considered one of the major limiting factors for crop production and contribute significantly to yield losses29,30,31. In addition to the competition with the growing crop in the field, they also act as reservoirs in the field for crop diseases and other insect pests30. On average, the losses due to the weeds amount to billions of dollars annually32. Manual eradication of weeds is a labor-intensive, time-consuming, and less efficient process29. Biotechnology offers exciting opportunities for the modification of crops with an intrinsic herbicide resistance make-up without instigating environmental complications33,34,35.
Event characterization of transgenic plants is the requirement for claiming intellectual property rights (IPRs) and meeting other regulatory requirements for the commercialization of the product36,37. It is done by identifying the unknown DNA sequences from the genome flanking the transgene cassette38. Different techniques have been used for this purpose in the past including genome walking (GW)38 and Southern blotting (SB)39. GW and SB are the most widely applied methods for event characterization of GM events, but these are laborious, time-consuming, and expensive. GW in combination with the sequencing approach has also been used for the identification of transgenic events40,41. In the past few years, high throughput next-generation sequencing (NGS) technology has emerged as the most promising and efficient approach for the molecular characterization of transgene events. As compared to GW and SB, NGS has been found more sensitive to detecting small insertion/deletions and multiple transgene events in GM crops and other organisms42,43,44,45.
The objectives of this study were to develop triple gene cotton lines harboring insect and herbicide-resistance genes in one T-DNA and evaluate the spatio-temporal expression of cry1Ac, cry2Ab, and EPSPS genes in different parts of the plants including leaves, bolls, and seeds at different growth stages. Another objective was to check the efficacy of Bt toxins, through insect bioassays with H. armigera and S. litura, and herbicide tolerance through glyphosate assays using recommended (1900 mL/acre) dose. Another objective was to characterize the transgene integration event of the best-performing line using high throughput NGS technology. Here we have shown that the transgenes are stably expressed under field conditions and can be transferred together in elite local cultivars by a single cross.
Results
Construct development
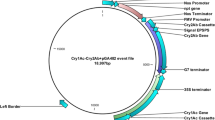
EPSPS cassette was successfully cloned in pGA482 followed by the cloning of two cassettes of Cry1Ac and Cry2Ab. This final vector pGA482-12ER encompassed triple gene cassettes including Cry1Ac (2 × 35S-Cry1Ac-35S), Cry2Ab (FMV-signal EPSPS-Cry2Ab-G7), and EPSPS (CVM-EPSPS-E9) as shown as vector map (Fig. 1). The sizes of Cry1Ac, Cry2Ab, and EPSPS cassettes were 3.5, 2.8, and 2.6 kb, respectively. The orientation of the cassette in pGA482 was confirmed through restriction analysis (Supplementary Fig. 1). The triple gene construct was named as pGA482-EPSPS-Cry2Ab-Cry1Ac (pGA482-12ER) and was successfully transformed. A confirmed clone of pGA482-12ER was transformed into Agrobacterium tumefaciens strain LBA4404 followed by cotton Coker312 transformation. The sequence of the gene construct has been submitted to GenBank under accession number KX880509.
Vector map of triple gene NIBGE cotton. Cry1Ac and Cry2Ab genes were to provide protection against chewing insect pests mainly bollworms and the EPSPS gene was for herbicide tolerance. Cry1Ac gene was cloned under 2X 35 promoter; Cry2Ab gene was cloned under FMV promoter and EPSPS was cloned under CVM promoter.
Development of transgenic Coker312 cotton harboring triple gene construct
Coker312 hypocotyls transformed with pGA482-12ER plasmid successfully started callus formation that led to the development of mature embryos. From mature embryos, T0 transgenic plantlets were successfully grown and shifted to the pots in greenhouse conditions. Various T0 transgenic lines were developed, and their molecular analysis confirmed the integration of triple gene construct in transgenic plants. After boll setting on T0 plants, seeds were harvested from these T0 transgenic lines and were further grown to achieve T1 generation. These T1 generation plants were then grown and NIBGE-E2, NIBGE-E3, and NIBGE-E20 cotton lines were selected for molecular analysis and insect bioassays.
Transgene analysis of T1 triple gene NIBGE cotton plants
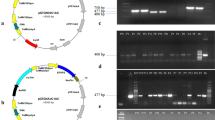
Putative transgenic plants of NIBGE triple gene cotton gave amplification of 521, 614, and 510 bp fragments corresponding to Cry1Ac, Cry2Ab, and EPSPS genes, respectively (Supplementary Fig. 2). Expression of the Cry1Ac, Cry2Ab, and EPSPS protein was successfully checked through the immunostrip and ELISA in all the three cotton lines of NIBGE triple gene cotton plants (Supplementary Fig. 3).
Quantification of Cry2Ab and EPSPS gene expression in T1 cotton plants
ELISA of NIBGE cotton lines was performed and gene expression was found comparable to the Monsanto cotton event. For the evaluation of spatial and temporal expression leaves, bolls, and seeds of NIBGE triple gene transgenic cotton lines were used to perform an ELISA assay. Inter and intra-plant variations of Cry2Ab expression was observed in NIBGE triple gene cotton plants. Maximum expression was found in leaves of 30 days old plants with a gradual decrease in the expression with the increasing age of the plant. The results have shown variable expression of Cry2Ab in different parts of the plant. Maximum expression was found in the leaves ranging from 7.3 to 8.77 µg/g of the tissue (Table 1), followed by maturing seeds 4.14–5.56 µg/g of the tissue, and the least expression was observed in boll rinds ranging from 1.7 to 2.7 µg/g of the tissue (Table 2).
Insect bioassay of T1 triple gene cotton plants
Positive transgenic plants were selected through immunostrip and PCR for insect bioassay with armyworm (Spodoptera litura). Significant mortality was observed in positive transgenic lines as compared to the non-transgenic Coker312 plants. Insect bioassay of these cotton lines showed 87 to 100% insect mortality using armyworm (S. litura). NIBGE-E2, NIBGE-E3, and NIBGE-E20 cotton lines showed 100, 89 and 87% mortality after 96 h, respectively (Fig. 2). Insect mortality data showed variations among the NIBGE triple gene cotton lines as statistically analyzed by ANOVA (p < 0.000). Similarly, the efficacy of triple gene NIBGE cotton against pink bollworm was successfully determined, Data was recorded after 21 days, and bolls were cut to count the number of survivors. NIBGE-E2 and NIBGE-E3 events showed 100% mortality while NIBGE E-20 event showed 86% mortality (Supplementary Figs. 4 and 5). Insect mortality data was in correspondence with gene expression levels of triple gene NIBGE cotton which was determined by ELISA (Fig. 2; Supplementary Figs. 4 and 5). Insect bioassay data were statistically analyzed by ANOVA.
Glyphosate assay
Immunostrip assay showed successful expression of EPSPS in NIBGE triple gene cotton plants, which were then quantified using ELISA. Maximum expression was found in NIBGE-E2 cotton line, followed by NIBGE-E3 and NIBGE-E20. Whole plant glyphosate assays of triple gene cotton showed promising results. NIBGE-E2 showed maximum tolerance against herbicide Galaxy at 1100 mL/acre dose at 35 days old plants. NIBGE-E3 and NIBGE-E20 had shown tolerance against herbicide Galaxy at 900 mL/acre dose. The efficacy of glyphosate assay was consonant with the expression of EPSPS gene in NIBGE triple gene cotton. NIBGE-E3 and NIBGE-E20 cotton lines did not tolerate the 1900 mL/Acre dose but survived on a 900 mL/acre dose. NIBGE-E2 cotton line exhibited significant tolerance to herbicide Galaxy even at 1100 mL/Acre dose (Fig. 3).
NGS-based event characterization
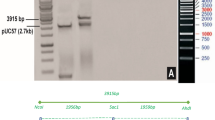
Whole genome sequencing of NIBGE-E2 cottonresulted in 648.37 M (97.26 Gb) raw bases. After removing low-quality reads, we obtained 638.44 Mb clean reads (95.77 Gb) with a 98.47% clean ratio. The clean reads of the sample had high Q20 (97.73%) and Q30 (92.67%), which showed high sequencing quality. The average GC content was 35.43%. The summary of sequencing results has been shown in (Supplementary Tables 1 and 2). Total clean reads per sample were aligned to the reference genome (Gossypium_hirsutum/HAU-AD1_genome_v1.0_v1.1) using Burrows-Wheeler Aligner (BWA) (Li 2013). The reference genome size is about 2.35 Gb (https://www.cottongen.org/species/Gossypium_hirsutum/HAU-AD1_genome_v1.0_v1.1), contains 2190 scaffold, GC content is 34.3%. The alignment showed that on average, 99.84% mapped successfully and 87.61% mapped uniquely. The duplicate reads were removed from total mapped reads, resulting in about 1.79% duplicate rate and 40.97-fold mean sequencing depth on the whole genome excluding gap regions. We obtained 97.66% of the whole genome excluding gap regions that were covered by at least 1X coverage, 96.68% had at least 4X coverage and 95.34% had at least 10X coverage. In addition, the distributions of per-base sequencing depth and cumulative sequencing depth are shown in (Supplementary Figs. 6–8), respectively. The insert size distribution of paired sequencing reads was plotted (Supplementary Fig. 9). The whole genome sequence of NIBGE triple gene cotton was successfully matched to the NIBGE triple gene construct and flanking regions were identified which was designated as the NIBGE-20-01 cotton event (Supplementary File; Additional Information). Furthermore, the event-specific primers designed for this region detected the specific amplicons as depicted in Table 3 which validated the NIBGE-20-01 cotton event (Supplementary Figs. 10 and 11).
Field assessment of triple gene NIBGE cotton
Each positive plant for event-specific primers was found to have intact three genes (Cry1Ac, Cry2Ab, and EPSPS) in a single cassette indicating single locus inheritance of the multi-transgenic cassette and proving that multi-transgene cassette can be deployed into elite cotton cultivars instead of single gene transfer. In field conditions, triple gene NIBGE cotton provides significant protection against the pink bollworm and other chewing insect pests including H. armigera and S. litura. Triple gene NIBGE cotton crossed with FH-1000 (Non-Bt), Bt-30 and Bt-85 showed 100% protection against the chewing insect pests as compared to NIA-Bt-1, NIA-88 (non-Bt. control), and Sohni (Non-Bt check variety) where pink bollworm infestation was observed to be 4.76%, 5.52%, and 7.31%, respectively (Supplementary Table 3).
Discussion
Genetic engineering provides an opportunity to modify crops to meet the world’s growing challenges of food security and environment-friendly strategies for controlling devastating insect pests. Cotton is one of the most widely cultivated crops in the world with a huge impact on the economy, especially in developing countries. Bt cotton was initially introduced in 1996 containing the Cry1Ac gene, providing resistance against the chewing insect pests and revolutionized pest control entailing a significant increase in the yields. However, in recent years pink bollworm has developed resistance against Cry1Ac protein. We identified mutations in the cadherin gene of pink bollworm that are likely to be involved in the resistance development (data not shown). Moreover, we showed that transgenic cotton plants expressing both Cry1Ac and Cry2Ab are resistant to pink bollworm under field and glasshouse conditions (data not shown).
In this study, we have designed a triple gene construct containing Cry1Ac and Cry2Ab genes for providing resistance against the chewing insect pests and EPSPS gene for herbicide tolerance. All three genes are cloned in the same T-DNA5 under different promoters and were transformed into cotton. This strategy was developed to help breeders in developing new cotton cultivars by incorporating these genes into the non-transgenic cotton cultivars or cotton cultivars containing a single Bt gene Cry1Ac (Mon531), where all three genes are inherited together. Previously, transgenic cotton with herbicide and insect resistance cotton varieties have been developed but those were developed through traditional breeding techniques between the herbicide tolerant and insect resistant cotton varieties.
To control the chewing insect pests, especially bollworms it is highly indispensable to have a sustainable expression of Bt toxins Cry1Ac and Cry2Ab in the vulnerable parts of the plant. It has been shown that the level of toxin declines with the age of the plant and even drops below the critical level at later stages which imposes high selection pressure on the insect pests to develop resistance against toxins24,46,47. In this study, we have quantified the spatio-temporal expression levels of the toxin in leaves, maturing seeds, and boll rinds of the green bolls which are considered as the most vulnerable parts of the plant, especially bollworms. The highest level of the toxin was found in leaves at the seedling stage followed by the maturing seeds and the lowest level of the toxin was found in the boll rinds of green bolls which shows variations throughout the growing season of the plant and was found variable among the triple gene cotton events (NIBGE-E2, NIBGE-E3, and NIBGE-E20).
Apart from the insect pests, weeds are another area that have gained significance due to the higher cost of labor for manual weeding and climate change that is resulting in unpredictable rain patterns in cotton growing areas. Therefore, cause significant loss in the crop yield resulting in decline yield. In the past weeds culminated through mechanical, cultural, and biological means which is a difficult and time-consuming process32,33,35. The discovery of glyphosate and its use in mid-nineties contributed vastly to the increase of world food production. In this study, a triple gene cotton containing EPSPS gene along with pest resistance Bt toxin genes Cry1Ac and Cry2Ab have been developed. The expression of the EPSPS gene in triple gene NIBGE cotton events was determined. The highest expression of the EPSPS gene was found in the NIBGE-E2 event while the lowest in the NIBGE-E20 event. Efficacy of EPSPS gene was confirmed through glyphosate assay at seedling stage and NIBGE triple gene cotton event E2 have shown maximum tolerance against the herbicide Galaxy @1100 mL/acre dose. E3 and E20 NIBGE triple gene cotton events were mildly tolerant to herbicide Galaxy @1100 mL/acre dose and were fully tolerant @900 mL/acre dose.
Our objective was to develop triple gene cotton containing insect and herbicide resistance genes under different promoters for a sustained expression of genes and in a single T-DNA which will be helpful for breeders for introgression in elite cultivars. Bollworms, H. armigera, and S. litura mainly feed on seeds more preferably than foliage and consistent expression of Cry1Ac is necessary for the developing seeds6. We selected the best performing event and used the whole genome sequencing approach for event characterization41,42,43,44,45 and the development of PCR based event-specific diagnostic tool. We showed that the diagnostics can distinguish NIBGE events from previously reported events. These diagnostics will not only help in securing intellectual property rights but also help breeders in the selection of desirable plants in the selection process. In the future tissue-specific promoter like alpha globulin and nodulin-like promoters may also be used for a sustainable expression of Bt toxins in seeds and reproductive parts, respectively. Apart from transgenic technology where two Bt genes are expressed together, other practices like mating disruptions or sterile insects must be used for effective and sustainable control of pink bollworms and other bollworms48,49,50,51.
Material and methods
Triple gene (Cry1Ac + Cry2Ab + EPSPS) construct development
All three genes (Cry1Ac, Cry2Ab, and EPSPS) were codon optimized for the cotton genome and were commercially synthesized. EPSPS full cassette (about 2.6 kb) was restricted with XbaI (Thermo Fisher Scientific ER0682) and HindIII (Thermo Fisher Scientific ER0502) from EPSPS cassette cloned in pBlue vector5. Binary vector pGA482 was also restricted using the pair of XbaI (Thermo Fisher Scientific ER0682) and HindIII (Thermo Fisher Scientific ER0502) and was ligated to EPSPS full cassette forming the clone named EPSPS-pGA482. EPSPS-pGA482 was restricted with HpaI (Thermo Fisher Scientific ER1031; blunt end cutter) and ligated to SwaI (Thermo Fischer Scientific ER1241) restricted double gene (Cry1Ac + Cry2Ab) construct that was already developed as described by Siddiqui et al.6. After screening, a positive clone was named pGA482-12ER, and orientations of genes were verified by restriction digestion.
Cotton transformation
Triple gene vector, pGA482-12ER having Cry1Ac-Cry2Ab-EPSPS cassettes was transformed into Agrobacterium strain LBA4404, and clones were confirmed through colony PCR. The confirmed clone was further used for the transformation of cotton var. Coker312. Hypocotyls were excised from 7 days old plantlets of Coker312 and were immersed into Agrobacterium culture of the pGA482-12ER plasmid. These hypocotyl cuttings were dipped in bacterial culture and were occasionally shaken. Afterward explants were dried on a blotting paper, followed by their transfer onto co-cultivation MS medium (MSP09-100LT, Caisson Labs-USA), where for two days; these were incubated in dark at 26 ± 2 °C. Then for callus initiation, these hypocotyls were further shifted to the callus induction medium. After callus formation, the calli were placed on an embryo maturation medium. Once embryos started maturing, these were shifted to germination medium followed by shifting onto shooting and rooting media in jars. Throughout the process, the shooting and rooting medium was supplemented with 50 mg/L kanamycin (Carl ROTH) and 200 mg/L cefotaxime. The plantlets with fully established roots were shifted to pots containing peat moss and then after hardening was shifted to pots having soil.
Molecular analysis of transgenic cotton plants
Molecular analysis of putative transgenic cotton plants was made through immunostrip test and polymerase chain reaction (PCR). For the immunostrip test (Catalog No. AS-046-LS, Envirologix USA), 20–25 mg of leaf tissue was brought to lab in a 1.5 mL microcentrifuge tube and placed on ice. Leaf was ground in 300 µL of extraction buffer given in the kit (Catalog No. AS-046-LS, Envirologix USA) using mortar and pestle and then centrifuged at 13,000 rpm for one minute. The supernatant containing protein was collected in a separate sterile 1.5 mL microcentrifuge tube, and an immunostrip for the triple gene (Cry1Ac, Cry2Ab, and EPSPS) was inserted in the supernatant and allowed to stay at room temperature to develop the required bands. Positive plants were selected and further evaluated through standard PCR using gene-specific primers (Table 4). DNA was extracted from the positive transgenic plants through CTAB (Cetyl Trimethyl Ammonium Bromide) method52. For PCR amplification, NIBGE gene-specific primers for Cry1Ac, Cry2Ab, and EPSPS were used. Monsanto event-specific primers for Bollgard II and Mon531 were used to rule out the presence of Monsanto events in NIBGE triple gene cotton lines. SadI primers6 were used as an internal control to check the quality of DNA. PCR reaction mixture was prepared using 7.5 µL Dream Taq Green Master Mix (Cat No. K1081, Thermo Fisher Scientific), 5 µL (50 ng/µL) genomic DNA from cotton, 1 µL (10 µM) of each gene, and event-specific primer53 and 11.5 µL of deionized water was used to make total reaction volume of 25 µL. A Bio-Rad PCR thermal cycler (C1000 Touch™) was used with the following PCR profile in which initial denaturation was done at 95 °C for 5 min, then 40 cycles with denaturation at 95 °C for 40 s, annealing at 55 °C for 40 s and then extension at 72 °C for 1 min, and with a final extension of 10 min at 72 °C. Primer sequences for nptII, Sad1, MON15985, MON531, and MON1445 were retrieved from the GMO Detection Method Database (GMDD; www.gmdd.shgmo.org) and the published literature53.
Quantification of Bt gene Cry2Ab
Spatio-temporal expression of Cry2Ab was quantified through ELISA (enzyme-linked immunosorbent assay) using an ELISA kit (Catalog No. AP 005, Envirologix USA). Ten seeds from each of three triple gene cotton lines (NIBGE-E2, NIBGE-E3, and NIBGE-E20) were grown in the greenhouse under controlled conditions (temperature, humidity) along with controls including Coker312 as negative control and Monsanto event Bollgard II (Mon15985) as a comparative control. After the identification of positive transgenic cotton plants through immunostrip and PCR analysis, ELISA was performed on different parts of the plant like the leaf, boll rind, and seeds. 10–15 mg of tissue was taken in a 1.5 mL microcentrifuge tube submerged in ice was taken and brought to the lab. ELISA was performed according to the given protocol with the kit (Cat No. AP 005, Envirologix, USA) and Cry2Ab expression was quantified and shown in µg/g of tissue weight by drawing a standard curve that was drawn using OD values from calibrators.
Glyphosate assay
Glyphosate assay of NIBGE triple gene cotton lines was performed at the seedling stage by using different doses of herbicide Galaxy i.e., @1900, 1500, and 1100 mL/acre dose. The herbicide solution was prepared and sprayed on thirty days old plants carefully. Coker312 and wild grass were used as negative controls and the equal dose was also sprayed on Coker312 and wild grass. The experiment was conducted under controlled conditions in a greenhouse and the data was recorded after 14 days of spraying.
Insect bioassays of transgenic cotton
To evaluate the efficacy of transgenic cotton lines insect bioassay was conducted with armyworm (S. litura). Three transgenic cotton lines (NIBGE-E2, NIBGE-E3, and NIBGE-E20) were used to check the efficacy of Bt toxins (Cry1Ac and Cry2Ab). Coker312 was used as a negative control and Bollgard II cotton (Mon15985) was used as a positive control in this experiment. Five plants of each line were selected randomly and three leaves from each plant were taken to be used in the bioassay. Each leaf was placed on a plate on filter paper and the leaf petiole was wrapped with wet cotton to keep the leaf fresh. Each leaf was exposed to five first instar larvae. During the experiment temperature and relative humidity were maintained between 25 ± 1 and 50–70%, respectively. Insect bioassay was performed twice with the same lines and controls entailing non-transgenic Coker312 as a negative control and Bollgard II cotton (Mon15985) as a positive control. Data was recorded to count the mortality rate of larvae every 24 h for five consecutive days until the completion of the bioassay. For calculating the difference in mortality between the transgenic and non-transgenic cotton lines, data was properly analyzed statistically using the analysis of variance (ANOVA) and least significant difference test (LSD).
Boll bioassay against pink bollworm
Boll bioassay of triple gene NIBGE cotton was conducted at NIBGE. Seeds of triple gene NIBGE cotton events (NIBGE-E02, NIBGE-E03, and NIBGE-E20) were grown in a greenhouse under controlled conditions. Once germinated, transgenic plants were verified with immunostrip (Catalog No. AS-046-LS, Envirologix, USA) and PCR using gene-specific primers for the Bt genes (Cry1Ac and Cry2Ab). Plants were grown and maintained in the greenhouse of NIBGE, without applying any insecticide. Ten days old bolls were collected from the plants and brought to the insect molecular biology lab of NIBGE. Bolls were briefly washed with running water and blot dry. Five bolls from each event were placed in a container. The container was closed with cloth lining to increase the ventilation and to control the fungal growth due to moisture as much as possible. Fifteen first instar larvae were carefully placed in each container using a fine brush. Bolls from the non-Bt cotton (Coker-312) and BGII (Mon15985) were used as a negative and positive control, respectively. After 22 days, all bolls from Bt (triple gene NIBGE cotton & BGII) and non-Bt (Coker312) were cut, and a number of survivors were counted inside the boll. The survival rate was calculated as the number of survivors divided by the total number of larvae placed in the container.
NGS-based event characterization of triple gene NIBGE cotton
Based on the gene expression and bioassays, the best performing transgenic cotton line (NIBGE-E2) was selected and further used for event characterization using NGS technology. Highly purified good quality genomic DNA was isolated from confirmed -triple gene NIBGE-E2 cotton plant by using PureLink™ genomic plant DNA purification kit (Catalog No. K183001, Thermo Fisher Scientific) and single replicate of NIBGE-E2 cotton was sequenced on BGISEQ-500 platform. Sequencing data in the form of paired-end reads stored in FASTQ format was obtained and data filtering was done to remove adapters, noisy or low-quality base ratio reads, and reads with unknown 'N' bases. The cleaned data were aligned to the reference cotton genome using Burrows-Wheeler Aligner (BWA) (Li 2013); followed by alignment with the triple gene NIBGE cotton construct sequence. The alignment was performed in CLC Genomics Workbench to detect the event in the genome of NIBGE triple gene cotton (. For NGS-based detected events in triple gene cotton, the event-specific primers (Supplementary Table 2) were designed from the junction region of the cotton genome and triple gene construct to validate the results. Event-specific primers were thoroughly evaluated and confirmed to be different from Monsanto cotton BGI (Mon531) and BGII (Mon15985) events.
Field evaluation of NIBGE triple gene transgenic cotton lines
The research work was conducted at Plant Breeding and Genetics Division, Nuclear Institute of Agriculture (NIA), Tando Jam, Pakistan during the Kharif seasons of 2019–2020 and 2020–2021. The experimental material consisted of triple gene (NIBGE-20-01) cotton developed at NIBGE Faisalabad, Non-Bt and Bt controls, and different crosses which were developed at NIA, Tando Jam.
For crossing, unopened flowers commonly known as candles or buds were hand emasculated in the evening and covered with butter paper bags. Male parent flowers were also covered with butter paper bags to avoid insect-intervened pollen contamination. Pollination of flowers was carried out in the morning (10:00 AM) and pollinated flowers were again covered with butter paper bags. At maturity, seed cotton was picked from all cross combinations. Ginning was performed and F0 seeds were collected for evaluation in the F1 generation. During Kharif 2020-21, F1 genetic material along with controls was planted in field conditions. Each plant in the F1 generation was tested for the presence of a triple gene cassette using immunostrip assay (Catalog No. AS-046-LS, Envirologix, USA) and through PCR by using NIBGE triple gene cotton event specific primers. Data on single plants were recorded for pink bollworm infestation (%) and seed cotton yield (g/plant). The percent infestation by pink bollworm was estimated by collecting boll samples from plants. Green susceptible bolls (bolls that can be easily pressed between the index finger and thumb) were collected from each plant, dissected, and examined for the presence and damage caused by pink bollworm larvae. Two observations were recorded with 15 days intervals in peak infestation period i.e., during September.
Percentage infestation of pink bollworm was calculated by using the formula:
Ethical statement
The study was approved by the Institutional Biosafety Committee (IBC) of the National Institute for Biotechnology and Genetic Engineering. The approval was given under the number IBC No. NIBG-49-112. All genetic manipulation in plants is approved by Institutional Biosafety Committee, duly registered with National Biosafety Committee (NBC). The case NIBG-49-112 was recommended by Technical Advisory Committee and approved by NBC.
Handling of plants/seeds
Handling of plants/seeds were carried out by relevant guidelines and regulations.
Data availability
The SRA datasets generated and/or analyzed during the current study have been submitted to NCBI with the accession number PRJNA801917 and can be accessed through the following link https://www.ncbi.nlm.nih.gov/sra/PRJNA801917.
References
Division, F. Economic survey of Pakistan. (2017–2018).
Babar, T. K. et al. Performance of some transgenic cotton cultivars against insect pest complex, virus incidence and yield. Pak. J. Agric. Sci. 50, 1–10 (2013).
Mansoor, S., Zafar, Y. & Briddon, R. W. Geminivirus disease complexes: The threat is spreading. Trends Plant Sci. 11, 209–212 (2006).
Ali, A. & Aheer, G. Varietal resistance against sucking insect pests of cotton under Bahawalpur ecological conditions. J. Agric. Res. 45, 3 (2007).
Naqvi, R. Z. et al. Development of a triple gene Cry1Ac-Cry2Ab-EPSPS construct and its expression in Nicotiana benthamiana for insect resistance and herbicide tolerance in plants. Front. Plant Sci. 8, 55 (2017).
Roberts, A., Nazli, H., Wach, M. & Zafar, Y. (International Life Sciences Institute, 2012).
Siddiqui, H. A. et al. Development and evaluation of double gene transgenic cotton lines expressing Cry toxins for protection against chewing insect pests. Sci. Rep. 9, 11774. https://doi.org/10.1038/s41598-019-48188-z (2019).
Damalas, C. A. & Eleftherohorinos, I. G. Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Public Health 8, 1402–1419 (2011).
Gerhardson, B. Biological substitutes for pesticides. Trends Biotechnol. 20, 338–343 (2002).
Mann, R. & Kaufman, P. Natural product pesticides: Their development, delivery and use against insect vectors. Mini Rev. Org. Chem. 9, 185–202 (2012).
Naqqash, M. N., Gökçe, A., Bakhsh, A. & Salim, M. Insecticide resistance and its molecular basis in urban insect pests. Parasitol. Res. 115, 1363–1373 (2016).
Brief, I. Brief 49-2014: Executive Summary. Global Status of Commercialized Biotech/GM Crops (2014).
ISAAA. Global Status of Commercialized Biotech/GM Crops in 2018: Biotech Crops Continue to Help Meet the Challenges of Increased Population and Climate Change. (ISAAA, 2018).
Briefs, I. Global Status of Commercialized Biotech/GM Crops in 2017: Biotech Crop Adoption Surges as Economic Benefits Accumulate in 22 Years. (2017).
Gujar, G. et al. Helicoverpa armigera baseline susceptibility to Bacillus thuringiensis Cry toxins and resistance management for Bt cotton in India. J. Invertebr. Pathol. 95, 214–219 (2007).
Soberon, M. et al. Engineering modified Bt toxins to counter insect resistance. Science 318, 1640–1642 (2007).
Tabashnik, B. E., Dennehy, T. J. & Carrière, Y. Delayed resistance to transgenic cotton in pink bollworm. Proc. Natl. Acad. Sci. USA 102, 15389–15393 (2005).
Fabrick, J. A. et al. Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to Bt cotton in India. PLoS ONE 9, e97900 (2014).
Tabashnik, B. E., Brévault, T. & Carrière, Y. Insect resistance to Bt crops: Lessons from the first billion acres. Nat. Biotechnol. 31, 510 (2013).
Tabashnik, B. E. & Carrière, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 35, 926 (2017).
Tay, W. T. et al. Insect resistance to Bacillus thuringiensis toxin Cry2Ab is conferred by mutations in an ABC transporter subfamily A protein. PLoS Genet. 11, e1005534 (2015).
Downes, S., Parker, T. & Mahon, R. Incipient resistance of Helicoverpa punctigera to the Cry2Ab Bt toxin in Bollgard II® cotton. PLoS ONE 5, e12567 (2010).
Mahon, R., Olsen, K., Garsia, K. & Young, S. Resistance to Bacillus thuringiensis toxin Cry2Ab in a strain of Helicoverpa armigera (Lepidoptera: Noctuidae) in Australia. J. Econ. Entomol. 100, 894–902 (2007).
Ullah, I., Asif, M., Arslan, M. & Ashfaq, M. Temporal expression of Cry1Ab/c protein in Bt-cotton varieties, their efficacy against Helicoverpa armigera (Lepidoptera: Noctuidae) and population dynamics of sucking arthropods on them. Int. J. Agric. Biol. 16, 579–585 (2014).
Tabashnik, B. E., Liu, Y.-B., Finson, N., Masson, L. & Heckel, D. G. One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc. Natl. Acad. Sci. USA 94, 1640–1644 (1997).
Tabashnik, B. E. et al. Insect resistance to Bacillus thuringiensis: Uniform or diverse?. Philos. Trans. R. Soc. Lond. 353, 1751–1756 (1998).
Kruger, M., Van Rensburg, J. & Van den Berg, J. Perspective on the development of stem borer resistance to Bt maize and refuge compliance at the Vaalharts irrigation scheme in South Africa. Crop Prot. 28, 684–689 (2009).
Shelton, A. M., Tang, J. D., Roush, R. T., Metz, T. D. & Earle, E. D. Field tests on managing resistance to Bt-engineered plants. Nat. Biotechnol. 18, 339 (2000).
Awan, M. et al. Transformation of insect and herbicide resistance genes in cotton (Gossypium hirsutum L.). J. Agric. Sci. Technol. 17, 1–10 (2015).
Délye, C., Jasieniuk, M. & Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 29, 649–658 (2013).
Dhaliwal, G., Jindal, V. & Dhawan, A. Insect pest problems and crop losses: changing trends. Indian J. Ecol. 37, 1–7 (2010).
Chauhan, B. S., Singh, R. G. & Mahajan, G. Ecology and management of weeds under conservation agriculture: A review. Crop Prot. 38, 57–65 (2012).
Green, J. M. The benefits of herbicide-resistant crops. Pest Manag. Sci. 68, 1323–1331 (2012).
Duke, S. Biotechnology: herbicide-resistant crops. Encyclop. Agric. Food Syst. 2, 94–116 (2014).
Green, J. M. Current state of herbicides in herbicide-resistant crops. Pest Manag. Sci. 70, 1351–1357 (2014).
Bhat, S. & Srinivasan, S. Molecular and genetic analyses of transgenic plants: Considerations and approaches. Plant Sci. 163, 673–681 (2002).
Privalle, L. S. et al. Development of an agricultural biotechnology crop product: Testing from discovery to commercialization. J. Agric. Food Chem. 60, 10179–10187 (2012).
Fraiture, M.-A., Vandamme, J., Herman, P. & Roosens, N. H. Development and validation of an integrated DNA walking strategy to detect GMO expressing cry genes. BMC Biotechnol. 18, 40 (2018).
Southern, E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503–517 (1975).
Bartholomae, C. C., Glimm, H., von Kalle, C. & Schmidt, M. in Mobile Genetic Elements 255–265 (Springer, 2012).
Volpicella, M. et al. Genome walking by next generation sequencing approaches. Biology 1, 495–507 (2012).
Yang, L. et al. Characterization of GM events by insert knowledge adapted re-sequencing approaches. Sci. Rep. 3, 2839 (2013).
Pauwels, K. et al. Next-generation sequencing as a tool for the molecular characterisation and risk assessment of genetically modified plants: Added value or not?. Trends Food Sci. Technol. 45, 319–326 (2015).
Zhang, R. et al. Molecular characterization of transgene integration by next-generation sequencing in transgenic cattle. PLoS ONE 7, e50348 (2012).
Kovalic, D. et al. The use of next generation sequencing and junction sequence analysis bioinformatics to achieve molecular characterization of crops improved through modern biotechnology. Plant Genome 5, 149–163 (2012).
Kranthi, K. R. et al. Temporal and intra-plant variability of Cry1Ac expression in Bt-cotton and its influence on the survival of the cotton bollworm, Helicoverpa armigera (Hubner) (Noctuidae: Lepidoptera). Curr. Sci. Bangalore 89, 291 (2005).
Chandrashekar, K., Kumari, A., Kalia, V. & Gujar, G. T. Baseline susceptibility of the American bollworm, Helicoverpa armigera (Hübner) to Bacillus thuringiensis Berl var. kurstaki and its endotoxins in India. Curr. Sci. 1, 167–175 (2005).
Siddiqui, H. A., Harvey-Samuel, T. & Mansoor, S. Gene drive: A faster route to plant improvement. Trends Plant Sci. 26, 1204–1206 (2021).
Xu, X. et al. Toward a CRISPR-Cas9-based gene drive in the diamondback moth Plutella xylostella. CRISPR J. 5, 224–236 (2022).
Harvey-Samuel, T. et al. Engineered expression of the invertebrate-specific scorpion toxin AaHIT reduces adult longevity and female fecundity in the diamondback moth Plutella xylostella. Pest Manag. Sci. 77, 3154–3164 (2021).
Min, J., Smidler, A. L., Najjar, D. & Esvelt, K. M. Harnessing gene drive. J. Respons. Innov. 5, S40–S65 (2018).
Porebski, S., Bailey, L. G. & Baum, B. R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15 (1997).
Asif, M. et al. Development of event-specific detection method for identification of insect resistant NIBGE-1601 cotton harboring double gene Cry1Ac-Cry2Ab construct. Sci. Rep. 11, 1–9 (2021).
Acknowledgements
The authors would like to thank Punjab Agricultural Research Board (PARB) Pakistan for providing funding to conduct this research under Project No. 426.
Author information
Authors and Affiliations
Contributions
S.M. and Z.M. conceived and designed the current project, advised during project execution, S.M. worked with collaborators, and finalized the manuscript. Z.M. was involved in designing of transgenes. H.A.S., M.A., and R.Z.N. performed cloning of transgenes, expression analysis, statistical data analysis, and interpretation of results. H.A.S. wrote the manuscript. S.A., Z.M., and M.Ar. were involved in cotton transformation and development of T0 and T1 transgenic cotton lines. M.S. was involved in the insect bioassays studies. I.A., M.F., C.L., and X.L. were involved in silico studies during event characterization. S.A. and M.R. were involved in making crosses and field evaluation studies. M.A., Z.M., and S.M. revised the manuscript critically.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Siddiqui, H.A., Asad, S., Naqvi, R.Z. et al. Development and evaluation of triple gene transgenic cotton lines expressing three genes (Cry1Ac-Cry2Ab-EPSPS) for lepidopteran insect pests and herbicide tolerance. Sci Rep 12, 18422 (2022). https://doi.org/10.1038/s41598-022-22209-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22209-w
- Springer Nature Limited