Abstract
Transgenic technology played a crucial role in developing insect-resistant plants resulting in the reduced application of pesticides. This article reports the expression of two cry proteins (Cry3Bb1 and Cry3) in cotton for enhanced resistance against chewing insect pests. The aforementioned genes were synthetically developed and were cloned under appropriate regulatory sequences followed by transformation into Eagle-2 genotype (Gossypium hirsutum) of cotton through shoot apex-cut Agro-infiltration. The transgene integration was validated by polymerase chain reaction using primers flanking the aforementioned cry genes. Transgene expression was assessed by qRT-PCR using GADPH as a reference gene. The relative fold expression analyses revealed the highest expression of the transgene(s) in M1 plants, which is a 4.5-fold expression (Cry3 + Cry3Bb1) followed by M3 (fold expression, 3.0) (Cry3Bb1) and M2 (fold expression, 2.5) (Cry3) transformants of cotton. The confirmed transgenic plants were exposed to insect pests, pink bollworm (Pectinophora gossypiella), and army bollworm (Helicoverpa armigera). Bioassay results revealed that 60% mortality was observed against pink bollworm, and 75% mortality was observed against army bollworm in transgenic plants containing both Cry3Bb1 and Cry3 genes (M1 transgenic plants). In M2 transgenic plants containing only the Cry3Bb1 gene, the mortality was observed to be 40% in the pink bollworm population, whereas 45% mortality was observed in the army bollworm population. In the case of M3 transgenic plants containing single gene-Cry3, the mortality was 20% in the pink bollworm population, whereas 30% mortality was observed in the army bollworm population. Almost no mortality was observed in non-transgenic Eagle-2 control plants. Hence, the developed cotton transformants have improved resistance against chewing insect pests.
Similar content being viewed by others
Introduction
Cotton (G. hirsutum) is a major agro-industrial crop grown worldwide for oil and fiber purposes1. The production of cotton is negatively affected by both biotic and abiotic stresses1. Different studies suggested that insect pests and diseases bring about 15–30% yield losses, and sometimes these losses can reach up to 50% due to direct injury and plant disease transmission2. The cotton fields are attacked by insect pests such as pink bollworm and army bollworm. The intensive application of chemical pesticides results in pesticide resistance in cotton pests, as well as these are also destructive to the environment and human health1. Pesticide applications also have negative impacts on beneficial insects3. A soil-borne bacterium, Bacillus thuringiensis (Bt), produces various insecticidal proteins (cry toxins), which have been used widely against many pests and until now, numerous cry toxins have been transformed in cotton4. Bt cotton reduces the pest burden without harming human health and the environment. Globally, the (Bt) cry3 proteins are toxic to several agricultural pests of the order Coleoptera, Lepidoptera, and Diptera5.
Despite the proven substantial protective effects of transgenic Bt-cotton plants against insect attack, this technology still needs improvement6,7. For instance, gene pyramiding involves defense strategies against insect pests8. The overproduction or multi-gene expression involved in insect defense can be a good alternative to reduce the pest attack and the development of insect resistance to cry toxins8.
It is interesting to mention that cry3Bb1 and cry3 have been successfully reported in several other crops, such as maize9, poplar10, eggplant11, and tobacco12, but it is hardly seen that these genes have been transformed into cotton. These genes have been proven effective strategy against insect pests in the aforementioned crops.
The early Bt genes transformed into maize were Cry1Ab, Cry1Ac and Cry2A 13. Subsequently, Cry34/35Ab1 and Cry3Bb1 genes were transformed for the management of closely related pest species14. Earlier, the stacking of multiple genes is reported in various crops, for example, a maize variety expressing five and six Bt genes (Cry1Ab, eCry3.1Ab, Cry1Fa2, mCry3A, and Vip3Aa20) and (Cry2Ab2, Cry1F, Cry1A.105, Cry35Ab1, Cry34Ab1 and Cry3Bb1) respectively15. Cry3Bb1 is one of the most commonly used Bt toxins in GM maize, has good insecticidal activity against Colorado potato beetle (Leptinotarsa decemlineata) and even better activity against western corn rootworm (D. v. virgifera)16. In eastern North Dakota (United States), the total feeding injury and population level of western corn rootworm were the lowest on Cry3Bb1 + Cry34/35Ab1 hybrids than on Bt maize producing either Cry3Bb1 or Cry34/35Ab1 protein alone17. In another study, the Cry3Bb1 gene expressed in maize showed resistance against Ostrinia furnacalis18.
In this proposed study, cry3Bb1 and cry3 have been tried in cotton against pink bollworm and army bollworm and produced a substantial effect. This study provides a new insight into how these cry genes can contribute significantly against insect pests in cotton.
Results
Development of plant transformation vector harboring double Cry gene(s)
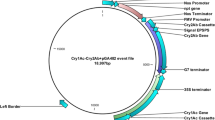
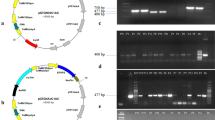
Two synthetic genes (Cry3Bb1 and Cry3) were synthesized from Synbio after codon optimization. Standard cloning was received in pUC57 cloning vector, wherein they were confirmed with AhdI, SacI and NcoI restriction endonucleases (Fig. 1a). The expected size of the cassette 3915 bp containing Cry genes was observed in Fig. 1a. A schematic diagram of the gene cassette carrying Cry3Bb1 and Cry3 genes is presented in Fig. 1b.
Restriction analysis of puc57 vectors containing the cry3Bb1 and cry3 gene cassette (total 3915 bp size) (A). NcoI and AhdI restriction analysis of the cry3Bb1 and cry3 gene cassette. M: 10 kb ladder. Lane 1: control of pUC57. Lane 2: cry3Bb1 and cry3 gene cassette. (B) Schematic representation of both gene cassette containing cry3Bb1 and cry3 genes.
The developed Cry gene(s) cassette was cloned in plant expression vector pCAMBIA1301 and was confirmed by restriction analysis with NcoI and AhdI (Fig. 2a). Cloning of the crystal protein genes (Cry3Bb1 and Cry3) was also confirmed by PCR (Fig. 2b).
Restriction digestion of both cry gene. (A) NcoI and AhdI restriction of pCAMBIA1301 vector containing cry genes cassette; M: 10 kb molecular weight marker, Lanes 1: shows blank, Lane 2: shows pCAMBIA1301, Lane 3: shows ligation without ligase, Lane 4, 5: shows ligation of cry genes with pCAMBIA1301. (B) PCR amplification of both cry genes. M: 10 kb molecular weight marker, Lanes 1, 2: show cry3Bb1 gene (459 bp), Lane 3, 5: show cry3 gene (562 bp), Lane 4: shows a blank.
Genetic transformation of cotton through Agrobacterium tumefaciens harboring double Cry gene(s)
The discrete colonies of A. tumefaciens strain LBA4404 transformed with crystal protein genes were randomly selected by colony PCR using gene-specific primers for cry3Bb1 and Cry3 genes. The PCR product was resolved on 1.5% agarose gel. The PCR product exhibited the expected size of the amplicon, 459 bp and 562 bp for Cry3Bb1 and Cry3 genes, respectively (Fig. 3).
The seeds of G. hirsutum variety Eagle-2 were delinted, surface sterilized using 5% HgCl2 and 1–2 drops of 2.5% SDS and incubated at 30 °C for 48 h. The germinated seedlings were used for transformation using the shoot apex cut method. The seedlings were incised and inoculated with A. tumefaciens cells containing both Cry3Bb1 and Cry3 genes (Table 1). The germination index and transformation efficiency of both control and transgenic plants were found to be 77.14% and 1.56%, respectively (Tables 2 and 3). A schematic transformation procedure is given in Fig. 4.
Molecular analyses of putative transformants of cotton
Fresh leaves were taken from the putative transformants of cotton. Total cellular DNA was isolated and was subjected to polymerase chain reaction using primers flanking the aforementioned cry genes. The PCR amplification resulted in product sizes of 459 bp and 562 bp for Cry (Cry3Bb1 & Cry3) genes, respectively (Fig. 5).
Total cellular mRNA was isolated from the leaves of putative cotton transformants and was used to synthesize cDNA. The resultant cDNA was used in order to track the expression of the transgenes (Cry3Bb1 and Cry3) using SYBR Green Supermix (Thermo Scientific, Cat#K0221) in the qRT-PCR assay. Relative expression analyses were carried out for the various transformants, whereas the GAPDH gene was used as an internal control for the normalization of reaction. Expression of both Cry genes was observed to be higher in M1 transgenic plants as compared to M2 and M3 transgenic plants, whereas no expression was detected in the non-transgenic cotton plants (Fig. 6).
Evaluation of transgenic cotton plants for resistance against insects.
The whole transgenic and non-transgenic plants were selected from the field for bioassay analysis. Transgenic plants (M1, M2 and M3), engineered with Cry3Bb1 and Cry3 genes, as well as non-transgenic plants, were exposed to pink bollworm and army bollworm. 5–7 3rd instar larvae were used in triplicates, and infestation data were recorded. On the third day of infection, 60% mortality was observed against pink bollworm, and 75% mortality was observed against army bollworm in transgenic plants containing both Cry3Bb1 and Cry3 (M1 transgenic plants). In M2 transgenic plants containing only Cry3Bb1, the mortality was observed to be 40% against pink bollworm, whereas 45% mortality was observed against army bollworm. In the case of M3 transgenic plants containing single gene-Cry3, the mortality was observed to be 20% against pink bollworm, whereas 30% mortality was observed against army bollworm. Almost no mortality was observed in non-transgenic Eagle-2 control plants (Fig. 7a,b).
(A) Cotton leaf mortality assay of army bollworm in different transgenic and non-transgenic cotton plants. Non-transgenic cotton plant leaf almost completely infected by army bollworm. (A) Transgenic cotton plant leaves containing both Bt-gene (cry3Bb1 and cry3), (B) shows transgenic plants containing cry3Bb1, (C) shows transgenic plants containing cry3, and (D) shows non-transgenic plants. (B) Pink bollworm mortality assay in transgenic and non-transgenic cotton plants. Non-transgenic cotton plant leaf almost completely infected by pink bollworm. (A) Transgenic cotton plant leaves containing both Bt-gene (cry3Bb1 and cry3), (B) shows transgenic plants containing cry3Bb1, (C) shows transgenic plants containing cry3, and (D) shows non-transgenic plants.
Two-way ANOVA showed the significance of our data at P ≤ 0.0001 (Fig. 8).
Discussion
Cotton being the leading fiber as well as an oil crop has great economic importance, and due to increased population pressure, its demand is also increased. So continuous improvement in the cotton crop against both biotic and abiotic stresses is important for cotton-growing countries. The invasive cotton pests like pink bollworm and army bollworm limit cotton production badly, so the transformation of novel genes is necessary for the development of resistance against these pests. For this purpose, a novel strategy was used; therein, a gene cassette was developed. The cassette contains two cry genes from B. thuringiensis. In the non-transgenic cotton variety Eagle-2, this cassette was transformed for the development of resistance against pests as reported in maize and tobacco. As reported earlier, the initial screening of putative cotton plants was performed with hygromycin19. These transgenic plants were confirmed by PCR as reported20. The qPCR was performed for PCR verified transgenic plants to determine mRNA expression21. Ultimately, to ascertain the effectiveness of transgene, an insect bioassay was performed against different target coleopteran and lepidopteran insects22.
In plants, several approaches are used for the insertion of a foreign gene, but Agrobacterium-mediated transformation is widely adopted in plant biotechnology. For instance, cotton shoot apex Agrobacterium-mediated transformation is more acceptable to exploit the transgenic technology in various cultivars23,24. From the early twentieth century, Agrobacterium and related species were known as plant pathogens. But with the continuous extensive research on Agrobacterium, it has been found that this bacterium can be used as a “natural genetic engineer”. Later studies showed that the involvement of host range, alterations in plant culture media and regeneration conditions are equally important along with improvement in a bacterium or host–pathogen25. The efficiency of Agrobacterium-mediated transformation depends on the type of promoter, Agrobacterium suspension medium, co-cultivation time and seed germination index26. Our study revealed a 77.14% seed germination index as well as 48% controlled and 1.56% experimental transformation efficiency.
Western corn rootworm is a serious threat to the maize crop in the USA. The upsurge in field evolved resistance against single Bt toxins reduced the efficacy of single gene transformation technology. Therefore, double or gene stacking technology is adopted to minimize the risks of resistance among various devastating pests. The transformation of the Cry3Bb1 gene is effective against western corn rootworms27. The transformation of both Cry3Bb and Cry3 in maize has produced many folds of higher expression against rootworms and leaf-feeding beetles5.
In this experiment, real-time qPCR assays were carried out to examine the mRNA expression of cry genes in transgenic plants. Compared to controls, the M1 plants revealed higher relative fold expression of 4.5 against pink bollworm and 4.3 against army bollworm than M2 (2.5 against pink bollworm and 2.2 army bollworm) and M3 (3.0 against pink bollworm and 2.5 against army bollworm).
Insect bioassays were conducted on the whole plant in the field. The data regarding infestation were recorded, and mortality of larvae was noted after three days in transgenic plants having both Bt Cry-genes, whilst no mortality was found in negative control cotton plants (Fig. 8). The larvae of pink bollworm and army bollworm were released on freshly growing leaves of the cotton plants. Transgenic cotton plants M1 exhibited 60% resistance towards pink bollworm whereas 75% resistance towards army bollworm for 3 days. In contrast, M2 transgenic plants revealed 40% and 45% resistance to pink bollworm and army bollworm. The transgenic plants M3 showed 20% and 30% resistance towards pink bollworm and army bollworm for 3 days. Our results suggested that stacking Bt Cry-genes is an efficient approach for managing various insect pests and minimizing the risk of resistance evolution in insects against transgenic cotton cultivars.
Materials and methods
Plant materials
The genotype Eagle-2 (Gossypium hirsutum L.) was used for the expression of two cry genes (cryBb1 and cry3). The healthy seeds of Eagle-2 were obtained from Four Brothers Seeds Multan-Pakistan and were grown at the research farm of Four Brothers Lahore-Pakistan.
Development of synthetic cry genes
Gene sequences of selected cry genes (cry3, accession no. AY572010.1; cry3Bb1, accession no. spIQ06117I1-652) were retrieved from NCBI and, after codon optimization, were synthesized by [BIOBASIC, CANADA]. The synthetic double cry3Bb1 gene and cry3 gene cassette (total 3915 bp) were cloned into the NcoI, SacI and Ahd1(restriction endonuleases) restriction sites of pUC57 vector. All genes were under regulation of CaMV35S promoter, and Nos terminator was added at the end of these genes (Fig. 1).
Development of plant transformation vector
The pUC57 vector carrying cry3Bb1 and cry3 genes cassette was transformed into the top 10 E. coli competent cells by using the heat shock method and selected on LB media containing ampicillin (50 µg/mL) and tetracycline (50 µg/mL). The Gene Jet plasmid DNA isolation kit (Thermo Scientific Vilnius, Lithuania, Cat#K0503) was used following the manufacturer’s instructions. For the confirmation of cry3Bb1and cry3 genes, restriction digestion was done by using NcoI and AhdI enzymes. A 3.9 Kb DNA fragment of cry3Bb1, and cry3 cassette were observed on 0.8% agarose gel. For the purification of eluted fragments, Gene JET Gel Extraction Kit (Thermo Scientific Vilnius, Lithuania, Cat#K0503) was used. The purified DNA fragment (cry3Bb1 and cry3 genes) was ligated into pCAMBIA1301 (plant expression vector), pre-digested with the corresponding restriction enzymes. A 10 Kb DNA marker (250 bp to 10 Kb) (GeneRuler, Thermo Scientific, Cat#SM0311) was used to assess the size of the resolved DNA fragments.
To confirm the ligation of the aforementioned cry genes in pCAMBIA1301 vector, restriction digestion was done with NcoI and AhdI enzymes. Further, cloning of these genes was also confirmed by PCR using gene(s) specific primers given in Table 4.
Following conditions were used for the amplification of cry genes; Initial denaturation (94 °C for 3 min), denaturation (94 °C for 45 s), annealing (59 °C for 50 s), extension (72 °C for 1:30 min) and final extension (72 °C for 10 min). By electroporation, the confirmed plasmids were transformed into A. tumefaciens strain LBA4404 competent cells28. The YEP media (Peptone 10 g/L, Yeast extract 10 g/L, Sodium chloride 5 g/L, pH 7.5) containing Kanamycin (50 mg/ml) and Rifampicin (50 mg/ml) were used for the selection of recombinant Agrobacterium cells. Colony PCR was performed for the selection of desired Agrobacterium clones.
Genetic transformation of selected cotton genotype
The surface sterilized seeds of Eagle-2 (upland cotton) were incubated in the dark at 30 °C for 48 h. The shoot apex cut method was used for the co-transformation of germinated seedlings. Before incubation, the isolated embryos were injured at the shoot apex and were treated with Agrobacterium strain LBA4404 harboring the pCAMBIA1301-cry. The cultures were incubated for 1 h. at 28 °C by placing explants on MS solid medium supplemented with (MS 4.4 g/L, Sucrose 30 g/L, Phytagel 2.4 g/L). After 48–72 h. of incubation, the embryos were cultured on MS media supplemented with cefotaxime (500 mg/ml), followed by screening in MS tubes containing hygromycin (25 mg/ml) for 1.5 months. After the screening, cotton plants were transferred into pots having an equal proportion of sand, clay and peat moss (1:1:1). Subsequently, the putative transgenic plants were shifted to greenhouses at Four Brothers Genetics Inc. for acclimatization and hardening. Thereafter molecular analyses were performed in order to assess for transgene integration and expression. The numerical data of genetic transformation was recorded in Table 1.
Detection of the double cry gene integration into cotton genome
Leaves (usually third leaf) were harvested from putative transgenic plants for the isolation of DNA for PCR-based screening of Cry3Bb1 and Cry3 gene(s) integration. For this purpose, the PCR master mix kit (Thermo Scientific, cat#K1081) was used with specific primers. To nullify the Agrobacterium contamination, the amplification of virG gene was also performed using a specific set of primers from the vir region.
Expression analysis of putative transformants of cotton
The Agilent kit (Agilent Technologies, Santa Clara, USA, Cat #5185-6000) was used for RNA isolation from putative cotton transformants. The Nano Drop ND-1000 spectrophotometer (at 260 and 280 nm) was used for the quantification of RNA. By using the first strand cDNA synthesis kit (Thermo Scientific, Cat #K1632), the cDNA was prepared from the DNase-treated total RNA. This cDNA was stored at −20 °C.
The qPCR was performed for expression analysis of transgenes with specific primers in triplicates with a Product size of < 200 bp using Maxima SYBR Green/ROX (Thermo Scientific, Cat#K0221). The reaction mixture of 20 µl with 1 µl of 10 pmol reverse and forward primers, 5 µl of Maxima SYBR Green/ROX qPCR Master Mix (2×) and 1 µl (50 ng/µl) of cDNA. The primer sequences for the amplification of both genes are given in Table 4. Relative expression was determined according to the 2 (−ΔΔCt) method using GAPDH primers as an internal control (reference gene) for normalization in reaction. All of the assays were carried out in triplicates.
Insect bioassays
Insect bioassay was executed to assess the toxic impacts of transgenic and non-transgenic cotton plants against larvae of pink bollworm and army bollworm. The expanded fresh leaves of PCR positive plants were identified and placed in a petri dish for bioassay. 5–7 larvae of army bollworm of 3rd instar were used per leaf in triplicate. Similarly, pink bollworms of the 3rd instar were released on the transgenic and non-transgenic plants in the field. The effectiveness of transgenic plants against pink bollworm and army bollworm was assessed. The two-way ANOVA was used for statistical analysis. For 3 days, the mortality rate was continuously recorded using the following equation;
Research involvement with human participants/ or animals
The research does not involve human and animal participants.
Informed consent
I have read and I understand the provided information and have granted permission to ask the questions. I understand my participation in the research and liable to produce the given information at any stage. The leaves of the plants were taken with the permission of FB Genetics Four Brothers Group Lahore.
Conclusion
The simultaneous expression of both Cry genes was assessed against various insect pests. The transgenic plants containing two Bt Cry-genes were compared with non-transgenic plants. The mortality of pink bollworm and army bollworm was observed considerable in transgenic plants harboring both Cry3Bb1 and Cry3 (M1) genes as compared to M2, M3 and negative control. The insertion of these Bt genes in cotton will attract the attention of the farmers who are walking out from the major cotton-producing countries around the world. The transformed genes used as a combining strategy, which are hardly seen in transgenic cotton plants, will help reduce the insect attack leading to the improvement in cotton yield.
Data availability
The datasets generated and/or analysed during the current study are available in the “[NCBI] repository (https://www.ncbi.nlm.nih.gov/search/all/?term=+AY572010.1)”.
References
Razzaq, A. et al. Pyramiding of cry toxins and methanol producing genes to increase insect resistance in cotton. GM Crops Food 12(1), 382–395 (2021).
McCook, S. & Vandermeer, J. J. P. The big rust and the red queen: Long-term perspectives on coffee rust research. Phytopathology 105(9), 1164–1173 (2015).
Siviter, H. & Muth, F. J. P. O. T. R. S. B. Do novel insecticides pose a threat to beneficial insects?. Proc. R. Soc. B 287(1935), 20201265 (2020).
Kamatham, S. et al. Recent advances in engineering crop plants for resistance to insect pests. Egypt J. Biol. Pest Control 31(1), 1–14 (2021).
Park, Y. et al. Enhancement of Bacillus thuringiensis Cry3Aa and Cry3Bb toxicities to coleopteran larvae by a toxin-binding fragment of an insect cadherin. Appl. Environ. Microbiol. 75(10), 3086–3092 (2009).
Kathage, J. & Qaim, M. J. P. O. T. N. A. O. S. Economic impacts and impact dynamics of Bt (Bacillus thuringiensis) cotton in India. Proc. Natl. Acad. Sci. 109(29), 11652–11656 (2012).
Kouser, S., Spielman, D. J. & Qaim, M. Transgenic cotton and farmers’ health in Pakistan. PLoS ONE 14(10), 222617 (2019).
Dorokhov, Y. L. et al. Metabolic methanol: Molecular pathways and physiological roles. Physiol. Rev. 95(2), 603–644 (2015).
Shrestha, R., Jakka, S. & Gassmann, A. J. J. O. A. E. Response of Cry3Bb1-resistant western corn rootworm (Coleoptera: Chrysomelidae) to Bt maize and soil insecticide. J. Appl. Entomol. 142(10), 937–946 (2018).
Génissel, A. et al. High tolerance against Chrysomela tremulae of transgenic poplar plants expressing a synthetic cry3Aa gene from Bacillus thuringiensis ssp tenebrionis. Mol. Breed. 11(2), 103–110 (2003).
Iannacone, R., Grieco, P. D. & Cellini, F. J. P. M. B. Specific sequence modifications of a cry3B endotoxin gene result in high levels of expression and insect resistance. Plant Mol. Biol. 34(3), 485–496 (1997).
Sutton, D. W., Havstad, P. K. & Kemp, J. D. J. T. R. Synthetic cryIIIA gene from Bacillus thuringiensis improved for high expression in plants. Transgenic Res. 1(5), 228–236 (1992).
Huang, F. et al. Inheritance of resistance to Bacillus thuringiensis toxin (Dipel ES) in the European corn borer. Science 284(5416), 965–967 (1999).
Masson, L. et al. A novel Bacillus thuringiensis (PS149B1) containing a Cry34Ab1/Cry35Ab1 binary toxin specific for the western corn rootworm Diabrotica virgifera virgifera LeConte forms ion channels in lipid membranes. Philos. Trans. R. Soc. 43(38), 12349–12357 (2004).
Xiao, Y. & Wu, K. J. P. T. O. T. R. S. B. Recent progress on the interaction between insects and Bacillus thuringiensis crops. Philos. R. Trans. Soc. 374(1767), 20180316 (2019).
Levine, S. L. et al. Independent action between DvSnf7 RNA and Cry3Bb1 protein in southern corn rootworm, Diabrotica undecimpunctata howardi and Colorado potato beetle, Leptinotarsa decemlineata. PLoS ONE 10(3), e0118622 (2015).
Calles-Torrez, V. et al. Transgenic Bt corn, soil insecticide, and insecticidal seed treatment effects on corn rootworm (Coleoptera: Chrysomelidae) beetle emergence, larval feeding injury, and corn yield in North Dakota. J. Econ. Entomol. 111(1), 348–360 (2018).
Kang, L. et al. Resistance to maize borer (Ostrinia furnacalis) of transgenic Bt maize and yield analysis. J. Maize Sci. 17(1), 62–70 (2009).
Singh, A. K. & Pental, D. Selection and genetic transformation of a fast-growing cell line in cotton (Gossypium hirsutum) for transgene expression studies. J. Plant Biochem. Biotechnol. 24(2), 225–232 (2015).
Yasmeen, A. et al. Amplicon-based RNA interference targeting V2 gene of cotton leaf curl Kokhran Virus-Burewala strain can provide resistance in transgenic cotton plants. Mol. Biotechnol. 58(12), 807–820 (2016).
Rao, A. et al. Variation in expression of phytochrome B gene in cotton (Gossypium hirsutum L.). (2013).
Khan, A. et al. Expression studies of chitinase gene in transgenic potato against Alternaria solani. Plant Cell Tissue Organ Cult. (PCTOC) 128(3), 563–576 (2017).
Guo, W.-F. et al. Rapid and convenient transformation of cotton (Gossypium hirsutum L.) using in planta shoot apex via glyphosate selection. J. Integr. Agric. 17(10), 2196–2203 (2018).
Zhang, B. Agrobacterium-mediated genetic transformation of cotton. In Transgenic Cotton 19–33 (Springer, 2019).
Gelvin, S. B. J. M. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67(1), 16–37 (2003).
Li, S. et al. Optimization of Agrobacterium-mediated transformation in soybean. J. Microbiol. 8, 246 (2017).
St. Clair, C.R., Head, G.P., & Gassmann, A.J.J.P.O. Western corn rootworm abundance, injury to corn, and resistance to Cry3Bb1 in the local landscape of previous problem fields. PLoS One 15(7), e237094 (2020).
Cristina, A., Maria, Q. & Vieira, M.-L. Plant transformation: Advances and perspectives. Sci. Agric. 56, 125 (1999).
Acknowledgements
I am grateful to The Institute of Cotton Research, Chinese Academy of Agricultural Sciences and FB Genetics Four Brothers Group for providing us with the expert opinion and facilities to perform the analysis and write the entire manuscript.
Funding
This work was supported by the Genetically Modified Organisms Breeding Major Project of China (2019ZX08010004-004) and the National Natural Science Foundation of China (31901579).
Author information
Authors and Affiliations
Contributions
M.M.Z.: Experimentation, Data collection, Drafting the manuscript; G.M.: Visualization, Validation, Review and editing manuscript; F.Sho.: Conceptualization, Resources, Supervision, Experimentation, Review, and Editing; A.I.: Formal Analysis, Visualization, Validation, Review, and Editing; A.S.: Experimentation, Data Acquisition, Review, and Editing; A.A.: Experimentation, Data Acquisition, Review and Editing; H.M.: Visualization, Validation, Review, and Editing; Y.Y.: Data Acquisition, Experimentation, Review, and Editing; A.R. and F.Sha.: Experimentation, Data Acquisition, Review, and Editing; M.R. and F.L.: Formal Analysis, Software, Visualization, Validation, Review, Editing, and Conceptualization, Funding, Supervision, Validation, Review, and Editing; All authors have reviewed the manuscript critically and approved the final draft for publication in Scientific report.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zafar, M.M., Mustafa, G., Shoukat, F. et al. Heterologous expression of cry3Bb1 and cry3 genes for enhanced resistance against insect pests in cotton. Sci Rep 12, 10878 (2022). https://doi.org/10.1038/s41598-022-13295-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-13295-x
- Springer Nature Limited
This article is cited by
-
Minimizing IP issues associated with gene constructs encoding the Bt toxin - a case study
BMC Biotechnology (2024)
-
Genome-wide identification and expression profiling of photosystem II (PsbX) gene family in upland cotton (Gossypium hirsutum L)
Journal of Cotton Research (2024)