Abstract
The forage species Caucasian clover (Trifolium ambiguum M. Bieb.), a groundcover plant, is resistant to both cold and drought. However, reference genes for qRT-PCR-based analysis of Caucasian clover are lacking. In this study, 12 reference genes were selected on the basis of transcriptomic data. These genes were used to determine the most stably expressed genes in various organs of Caucasian clover under cold, salt and drought stress for qRT-PCR-based analysis. Reference gene stability was analyzed by geNorm, NormFinder, BestKeeper, the ∆Ct method and RefFinder. Under salt stress, RCD1 and PPIL3 were the most stable reference genes in the leaves, and NLI1 and RCD1 were the most stable references genes in the roots. Under low-temperature stress, APA and EFTu-GTP were the most stable reference genes in the leaves, and the RCD1 and NLI2 genes were highly stable in the roots. Under 10% PEG-6000 stress, NLI1 and NLI2 were highly stable in the leaves, and RCD1 and PPIL3 were the most stable in the roots. Overall, RCD1 and NLI2 were the most stable reference genes in organs under normal conditions and across all samples. The most and least stable reference genes were validated by assessing their appropriateness for normalization via WRKY genes.
Similar content being viewed by others
Introduction
Caucasian clover (Trifolium ambiguum M. Bieb.), also known as Kura clover, is a high-quality rhizomatous perennial legume forage and groundcover species that originated in Russia and is distributed in areas with a cool and humid climate1. The dense rhizome system of Caucasian clover can store large amounts of metabolic energy that enables the perennial persistence and rapid reestablishment of this species2, and a unique characteristic of this species is the formation of daughter plants from the rhizomes that develop from the taproot. This characteristic confers an advantage to Caucasian clover under drought conditions3. To overcome abiotic stress (salt, cold, heat, and drought) and biotic stress (pests and pathogens), various mechanisms have evolved, but the molecular bases of these mechanisms are complex. A draft transcriptome of Caucasian clover has been assembled, mapped and functionally annotated and is available via public databases (accession number SRP159097 in the NCBI database). Nonetheless, few studies have focused on Caucasian clover gene expression. More research on the molecular mechanisms in Caucasian clover is needed.
The discovery of key genes involved in certain biological processes is the most important step in molecular research4. Several techniques are available to investigate gene expression, including semiquantitative reverse transcription polymerase chain reaction, northern blotting, in situ hybridization, and quantitative real-time PCR (qRT-PCR)5. A substantial amount of molecular research on plants has aimed to elucidate various processes through related functional genes. qRT-PCR is the most common technique used for validation of data from transcriptomic studies because of its reproducibility, rapidity, accuracy and sensitivity6.
Reference genes, also called housekeeping genes, are expressed stably in different tissues during different physiological states of organisms and in response to different environmental stimuli7. Ideal reference genes that are stably expressed during different biological and physiological states can be effectively used for normalization8. Several reference genes, including actin (ACT), elongation factor 1 alpha subunit (EF1α), glyceraldehyde-3-phosphase (GAPDH) and tubulin alpha (TUA), have been utilized for reliable qRT-PCR-based studies in various species9,10,11,12. Many reports have indicated that ideal stable reference genes do not exist and that stable reference genes differ among plant species, growth stages, growth conditions and treatments13. In addition, numerous studies have reported reference genes in various species, including Miscanthus lutarioriparia14, ladybird beetle15, pitaya16 and Hemarthria compressa17. Moreover, UBQ was found to be the most stable of seven candidate genes across all organs (leaves and stolons) and treatments (water-limited and well-watered conditions) in white clover (Trifolium repens L.)18, and Tr-β-ACTIN and Tr-GAPDH are two reference genes whose transcript abundance remains stable under biotic stress conditions in white clover19.
There are no reports in the literature on the evaluation of candidate reference genes for qRT-PCR in Caucasian clover. Thus, in this study, we selected 12 candidate reference genes: those encoding cell differentiation protein RCD1 homolog (RCD1)20, CYP, protein phosphatase type 2A complex (PP2A)21, NLI interacting factor-like phosphatase (NLI1), NLI interacting factor (NLI2), elongation factor Tu GTP-binding domain (EFTu-GTP), cationic amino acid transporter 9 (APA), RBD, protein tyrosine phosphatase activity (PTPMT1), tobamovirus multiplication-like protein (TMP), microtubule-associated protein CRIPT (MAP)21 and a bZIP transcription factor (BZIP). To investigate the most appropriate genes for normalization in this species and confirm the suitability of these genes, the relative expression levels of the target genes of WRKY were analyzed via qRT-PCR, with the most and least stable reference genes used. The results of this study will advance research on Caucasian clover.
Material and methods
Plant materials, growth conditions and abiotic stress treatments
Caucasian clover seedlings were used in this study. Original sources of the plant materials were obtained from the Inner Mongolia Grass Variety Engineering Technology Research Center of Inner Mongolia Agricultural University. Staff at the center formally identified the samples, provided details of specimen deposition and provided seeds (International Plant Name Index (IPNI) Life Sciences Identifier (LSID) urn:ipni.org:names:522843-1). After they were sterilized with 75% ethanol for 30 s, followed by NaClO4 for 10 min, the seeds were sown in vermiculite in plastic pots (10 cm diameter, 9 cm depth) in a greenhouse at Northeast Agricultural University on 9 September 2018. The greenhouse had a day/night average temperature of 24/18 °C, a photoperiod of 16/8 h (light/dark) and a relative humidity of 70–80%, and the vermiculite was kept moist with 1/2-strength Hoagland nutrient solution. The plants were treated when they were 5 weeks old. Samples from different organs (roots, stems and leaves) were collected from the plants for 5 weeks. The experimental design included three biological replicates.
Five treatments were applied in the present study: (1) a low-temperature treatment involving subjecting the plants to 4 °C for different durations (0, 2, 6, 12, and 24 h) under irrigation with Hoagland solution, after which leaf and root tissues were collected; (2) a salt treatment involving the application of 200 mL of NaCl at different concentrations (0, 12.5, 25, 50 and 100 mmol/L) to simulate high-salt conditions for 4 h, after which leaf and root tissues were collected; (3) a drought stress treatment involving subjecting plants to 10% polyethylene glycol (PEG)-6000 solution (w/v; Sangon, China) for different durations (0, 2, 6, 12, and 24 h), after which leaf and root tissues were collected; (4) a control in which stems, leaves and roots were collected under normal conditions; and (5) a treatment in which all samples from the four treatments were considered together for all samples. All the samples were collected in triplicate, frozen in liquid nitrogen, and stored at − 80 °C.
Total RNA isolation and first-strand cDNA synthesis
Total RNA was extracted from each sample by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Genomic DNA (gDNA) was removed by RNase-free DNase I digestion during the isolation procedure. The quantity and purity of RNA were determined via 1% agarose gel electrophoresis. RNA whose A260/280 ratio ranged from 1.8 to 2.2 and whose A260/230 ratio was greater than 2.0 was used for further synthesis. All the RNA samples were stored at − 80 °C. For qPCR, first-strand cDNA was synthesized from 1 μg of total RNA via HiScript II Q RT SuperMix for qPCR (+ gDNA wiper) according to the manufacturer’s protocol7. Before the next step, the cDNA samples were stored at − 20 °C.
Selection of candidate reference genes, mining of target transcripts, designing of primers and verification of selected gene amplicons
The RNA transcriptome sequence of Caucasian clover is available from a public database (accession number SRP159097 in the NCBI database). Twelve candidate reference genes among 27,004 genes (with fragments per kilobase of transcript per million mapped reads (FPKM) values ≥ 5 for at least three replicates per treatment and differences from each other not surpassing 5)22 identified via RNA sequencing analysis were selected from the RNA transcriptome sequence of Caucasian clover. In addition, data mining for target transcripts within the transcriptome was also performed.
The primer pairs of candidate reference genes and target transcripts were designed via the NCBI website (https://www.ncbi.nlm.nih.gov/) and Oligo 6 software, with the following parameters: a primer length of 20–22 bp (optimal length of 100 bp), an amplicon length of 80–150 bp, an annealing temperature (Tm) within the range of 57–63 °C (optimal temperature of 60 °C), and a temperature difference of each primer of less than 1 °C. All the primers used were synthesized by a commercial supplier (RiboBio, Harbin, China). All the information about the primer design in this study is listed in Table 1. Primer specificity was determined via 1.0% agarose gel electrophoresis (Fig. 1 and Fig. S1).
Specificity of Caucasian clover reference gene primer pairs for RT-qPCR amplification. Agarose gel (1.0%) electrophoresis displaying single PCR products with expected sizes for the 12 reference genes (right side, 50 bp DNA ladder). Figures 1–7 represent the RCD1, PPIL3, PP2A, NLI1, NLI2, EFTu-GTP and RBD reference genes, respectively, and the other reference genes identified via agarose gel electrophoresis are displayed in Fig. S1 of the additional files.
qRT-PCR and gene-specific PCR amplification efficiency
qRT-PCR was performed in 96-well plates on a Quantagene q225 Real-Time PCR System (Novogene). The total volume of the reaction solution was 10 μL, which consisted of 0.5 μL of cDNA, 0.2 μL of forward primer, 0.2 μL of reverse primer, 4.1 μL of ddH2O and 5 μL of 2× ChamQ Universal SYBR qPCR Master Mix. The PCR procedure was as follows: the thermal profile of the reaction included initial denaturation at 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s and 57 °C for 30 s and a final step of 72 °C for 15 s. A melting curve was produced after 40 cycles of amplification by heating at 65–95 °C to confirm the specificity of the PCR products. Slopes in the range of − 3.58 to − 3.10 were considered acceptable for the PCR assay.
The amplification efficiency E = 10(−1/slope of the standard curve) and R2 value for all primer pairs were determined from a 6-point standard curve produced by tenfold serial dilutions of cDNA, in triplicate23.
Ranking the stability of candidate reference genes
geNorm23, NormFinder24, BestKeeper25, the ΔCt method and RefFinder26 (https://www.heartcure.com.au/reffinder/?type=reference) were used to analyze the stability of the candidate reference genes under different conditions. All procedures were performed in accordance with the program instructions.
The geNorm algorithm, described by Vandesompele et al.27, calculates the gene expression stability, M, for a reference gene as the average pairwise variation, V, for that gene with all other tested reference genes. Stepwise exclusion of the gene with the greatest M value allows ranking of the tested genes according to their expression stability. Stable reference genes with M values lower than 1.5 can be used with geNorm to determine the optimal number of reference genes for normalization by the pairwise variation Vn/n+1. Variations with values below the threshold of 0.15 are considered ideal pairwise variations28,29.
NormFinder software was used to identify the stable reference genes, and the principle underlying the calculations was similar to that used by geNorm. First, we obtained the expression stability, M, and then selected the most stable reference gene according to M. The standard was the most stable reference gene with the smallest M value. However, this program selects only the most suitable reference gene.
The BestKeeper algorithm compares the expression levels of only ten reference genes and ten target genes in 100 samples. The correlation coefficient (r), standard deviation (SD) and coefficient of variation (CV) between each gene were obtained. The magnitudes of the respective values were then compared, after which the reference gene with the best stability was ultimately determined.
With the ΔCt method, ranks were determined according to pairwise comparisons of gene sets. The reference gene with the lowest SD had the most stable expression. RefFinder was then used to combine all four statistical methods (geNorm, NormFinder, BestKeeper and the ΔCt method) to calculate the comprehensive ranks.
Validation of reference gene stability
In the present study, the WRKY gene was verified to be involved in various processes, including responses to biotic and abiotic stresses. The relative expression data were calculated according to the 2−ΔΔCt method30 and presented as relative expression levels. The sequence of WRKY was obtained from the RNA sequence of Caucasian clover deposited in the NCBI database. In this study, we used WRKY as a target gene to validate the stability of the reference genes. The relative expression levels of WRKY in the roots of plants under low-temperature treatment and in the roots of plants under salt stress were determined and normalized by the use of the most and least stable reference genes.
Results
Primer verification and expression levels of candidate reference genes
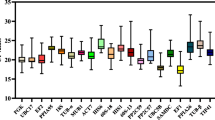
We determined the cycle threshold (Ct) values for 12 candidate reference genes (three biological replicates and three technical replicates). The Ct values of all the candidate reference genes are shown in Supplementary Table S1 and Fig. 2, and the expression levels of the candidate reference genes were measured. The Ct values ranged from 22.15 (PP2A) to 27.04 (NLI1). The variable Ct values of all the candidate reference genes among the different treatment conditions demonstrated that the expression levels varied on the basis of the conditions and experimental treatments. The expression of PP2A (Ct = 22.15), followed by that of PPIL3 (Ct = 24.55), RCD1 (Ct = 24.58) and NIL2 (Ct = 26.35), was little affected by the treatments, whereas the expression of NLI1 (Ct = 27.04) was strongly affected by the treatments. These results implied that the expression of all the reference genes was inconsistent across all the treatments and experimental conditions. Thus, it was necessary to select the most stable reference genes for normalizing gene expression in Caucasian clover under different conditions (Fig. 2).
Expression stability of candidate reference genes
To further evaluate the stability of the candidate reference genes, we used five methods (geNorm, NormFinder, BestKeeper, the ∆Ct method and RefFinder) to determine the individual expression stability of the genes.
geNorm analysis
In the geNorm analysis, the M values were determined for root and leaf samples of plants subjected to three different treatments (Fig. 3). The rank of the candidate reference genes differed on the basis of the different conditions. In the roots of plants under low temperature (Fig. 3A), PP2A and PPIL3 had the lowest M values, which indicated that they were most stable under the low-temperature treatment. NLI1 had the greatest M value (1.51), and its expression was the most unstable. In the leaves of plants under low temperature (Fig. 3B), all of the M values were below the threshold of 1.5, and the expression of PTPMT1 and MAP was the most stable. Overall, M is suggested to be the criterion for appropriate reference gene selection (Fig. 3). In the roots of plants under salt treatment, the M values of BZIP, RBD and MAP were > 1.5. In the leaves of plants under 10% PEG-6000 conditions, the M value of BZIP was > 1.5; all the other genes were perhaps more suitable reference genes for target gene normalization.
Average M values of the 12 candidate reference genes according to geNorm. The expression stability was evaluated in samples from organs, in all samples, and in the roots and leaves of Caucasian clover subjected to low-temperature, salt and 10% PEG treatments. The least stable reference genes, which have relatively high M values, are shown on the left, and the most stable reference genes, which have relatively low M values, are shown on the right.
We also calculated the optimal number of reference genes according to the pairwise variation results (Vn/n+1 values). The results showed that, with the exception of those of the roots of plants under salt stress (V4/5 = 0.13), all of the V2/3 pairwise variations calculated by geNorm were less than 0.15, and V4/5 was determined to be 0.13 for the roots of plants subjected to salt stress. These results suggested that the two most stable reference genes were adequate for qRT-PCR-based normalization across different experimental conditions and that an additional reference gene was not needed (Fig. 4).
Pairwise V values of 12 candidate reference genes from Caucasian clover, as calculated by geNorm. The different treatments are marked with square frames of different colors. LT indicates the low-temperature treatment (4 °C), and PEG indicates the 10% PEG-6000 treatment; all samples of the four treatments were considered jointly as all samples, and organs were evaluated under normal conditions, as described below.
NormFinder analysis
In NormFinder analyses (Table 2), relatively low stability values indicate relatively stable reference genes. The results of the NormFinder analysis are listed in Table 2. The most stable reference genes differed among organs and stresses. The most stable reference genes included RCD1 under salt stress, PPIL3 under 10% PEG-6000 stress and PTPMT1 under low-temperature stress across all the leaf samples. The most stable reference gene under salt and low-temperature stresses was PP2A, and that under 10% PEG-6000 stress was PTPMT1 across all the root samples. The most stable reference genes across all the organs and samples were EFTu-GTP and RBD, respectively. The least stable reference genes across all the samples and conditions were MAP and TMP, respectively.
∆Ct analysis
Relatively small ΔCt values of candidate reference genes indicate relatively high gene expression. As shown in Table 3, the ΔCt values differed among the stresses. In the organs, the smallest ΔCt values of the reference genes were 0.56, 0.56 and 0.58 for EFTu-GTP, RBD and NLI2, respectively, and the most unstable reference gene was TMP. However, the most stable reference genes were EFTu-GTP and NLI1 across all the samples, and PPIL3 was the most unstable reference gene overall. Among all the leaf samples, the most stable reference genes were PTPMT1 under low-temperature stress and PPIL3 under both salt stress and 10% PEG-6000 stress. Among all the root samples, the most stable reference genes were EFTu-GTP under low-temperature stress, PP2A under salt stress and MAP under 10% PEG-6000 stress.
BestKeeper analysis
In BestKeeper analysis (Table 4), the ranking of candidate reference genes is evaluated according to CV and SD values, which are determined by Ct values. Relatively high stability of reference genes is represented by relatively low SD and CV values, and candidate reference genes are unsuitable for normalization and unstably expressed when the SD is > 1.
As shown in Table 4, among all the leaf samples, the most stable reference genes were RCD1 under salt stress, PTPMT1 under 10% PEG-6000 stress and EFTu-GTP under low-temperature stress. In the leaf samples of plants treated with 10% PEG-6000, the SD values of NLI1 and BZIP were greater than 1, which indicated that the reference genes could not be used for normalization. Among all the root samples of the plants, the most stable reference genes were NLI2 under salt stress, PPIL3 under 10% PEG-6000 stress and PP2A under low-temperature stress. Among the different organs, the most stable reference gene was EFTu-GTP, and PTPMT1 was the most stable reference gene across all the samples. Candidate reference genes with an SD > 1 were identified in the roots of plants treated with salt, 10% PEG-6000 and low temperature; these genes could not be used for normalization.
RefFinder analysis
The online tool RefFinder was used to determine the comprehensive ranking of the candidate genes analyzed by the other four methods (geNorm, NormFinder, BestKeeper and the ΔCt method). The results of RefFinder also showed that the comprehensive rankings of the reference genes differed under the various stresses and among the different organs (Supplementary Table S2 and Table 5). The most stable reference genes were RCD1, NLI1 and APA in most of the samples under various stresses and in the different organs. In contrast, EFTu-GTP, BZIP, NLI2, PP2A and APA were the most unstable reference genes under various stresses and in different organs. Compared with the most unstable reference genes, the most stable reference genes were more consistent. We used RefFinder to calculate the geometric mean of the stability rankings obtained from the analysis of geNorm, NormFinder, BestKeeper and the ∆Ct method and obtained a comprehensive ranking (Supplementary Fig. S3).
Reference gene validation
To validate the ranking of the 12 candidate reference genes of Caucasian clover, the expression levels of the WRKY gene in the roots of plants under low-temperature treatment (Supplementary Table S3) and in the roots of plants under salt treatment (Supplementary Table S4) were normalized. WRKY proteins compose a large family of transcription factors involved in the abiotic stress response31, and many WRKY genes have been identified in Oryza sativa32, Hordeum vulgare33, Cucumis sativus34 and citrus species35.
On the basis of the comprehensive analysis of the results from geNorm, NormFinder, BestKeeper, and the ΔCt method by RefFinder, the two most stable reference genes and the least stable reference genes under different treatment conditions were selected. In root samples from plants subjected to salt and low-temperature treatments, the expression of WRKY at 0 h was assumed to be 1 and was used to compare the relative expression of genes from the samples at other time points.
As shown in Fig. 5, in the roots of plants under low-temperature stress, the two most stable reference genes, RCD1 and NLI2 (including RCD1 + NLI2), and the least stable reference gene, EFTu-GTP, according to the results of the comprehensive evaluation were used to calibrate the relative expression of the WRKY, RCD1, and NLI2 target genes, respectively. In addition to the expression pattern of the RCD1 + NLI2 combination, which was used as a reference gene, the expression pattern of WRKY was consistent. When the least stable reference gene (EFTu-GTP) was used to correct the target gene, the expression pattern greatly differed from the above expression pattern. In the roots of plants under salt stress, the relative expression of the WRKY target gene was validated using the two best reference genes, NLI1 and RCD1; the NLI1 + RCD1 combination; and the least stable reference gene, NLI2. When NLI1, RCD1, and the NLI1 + RCD1 combination were used as references, the expression pattern of WRKY was consistent. When the least stable reference gene (NLI2) was used to validate the target gene, the expression pattern was very different from the above expression pattern.
The relative expression level of WRKY was determined by the use of select reference genes, including the most or least stable reference genes, for normalization in the roots. (A) Relative quantification of WRKY expression in the roots of plants under low-temperature stress. (B) Relative quantification of WRKY expression in the roots of plants under salt stress. All the materials were treated for 0, 2, 6, 12 or 24 h. The error bars represent the standard errors of the means of three biological replicates. Statistical analysis was performed at different times for the same reference gene. The means with different letters are significantly different from one another (P < 0.01).
In the roots of plants under low-temperature treatment, the least stable reference gene was EFTu-GTP, and the most stable reference genes were RCD1 and NLI2. In the roots of plants under salt stress, the least stable reference gene was NLI2, and the most stable reference genes were NLI1 and RCD1. We also used the combination of the most stable reference genes for validation.
Discussion
As an advanced, accurate and commonly used research tool, qRT-PCR is pivotal for quantifying the relative expression levels of target genes36. qRT-PCR has been used to quantify relative levels of gene expression on the basis of normalization to the expression of stable reference genes37. Unstable reference genes can substantially affect the results of such analyses and can even lead to erroneous conclusions38. Primer amplification efficiency represents the amplicon doubling rate during the PCR process, which also affects the accuracy of qRT-PCR16. A good primer has an E value ranging from 90 to 105%23. In this study, the E values of all the primers used were in this range, ensuring the accuracy of the qRT-PCR data in this study.
After consulting previous studies of other species and searching our transcriptomic database, we selected 12 candidate reference genes for evaluation. No reference genes suitable for different tissues or organs of Caucasian clover related to growth and development and stress conditions were identified. In general, the most stable genes in all the samples were RCD1, PPIL3, NLI1 and NLI2, whereas bZIP and EFTu-GTP were the most unstable reference genes. bZIP and EFTu-GTP encode a transcription factor and an elongation factor Tu GTP-binding domain-containing protein, respectively. It is very easy to draw erroneous conclusions due to random effects caused by heterogeneity of the samples. Therefore, it is not recommended to use bZIP or EFTu-GTP as a reference gene in such experiments. RCD1 is needed to maintain cells in a division-competent state and to regulate division plane placement39. Reports on PPIL3, NLI1 and NLI2 in plants are scarce. PP2A was evaluated in four strains of Auricularia cornea40 at different developmental stages, and its comprehensive ranking was 3. The rankings generated from the four algorithms were incomplete, and the results were confirmed in Rhododendron20, Taihangia rupestris41 and Baphicacanthus cusia42. In the leaves and fruits of Lagenaria siceraria43, the reference gene CYP was the most unstable according to geNorm.
Nearly all of the relevant studies have revealed that the use of more than one reference gene for normalization yields more accurate results44,45. In most cases, the gene stability results calculated by geNorm and NormFinder were similar (Fig. 3 and Table 2). The best number of reference genes used for normalization was calculated by geNorm, and the results showed that, with the exception of the value from root samples of plants under salt stress, all the V2/3 values were below the threshold (0.15). A V2/3 value of < 0.15 indicated that the optimal number of reference genes for normalization was two, and a V4/5 value of < 0.15 in the roots of plants under salt stress indicated that the best number of reference genes for normalization was four, which was not ideal. However, the M values calculated by geNorm can also be used to evaluate the stability of reference genes; the results showed that only three of the studied reference genes were unstable, while the M values of the other reference genes were below 1.50. Thus, V scores cannot be used as the only index. On the basis of the results of this study, we suggested that using the most stable reference gene for normalization may be a better choice for the roots of plants under salt stress conditions. However, the gold standard of qRT-PCR is the use of at least four reference genes to determine the deviation of a single reference gene38. Using four algorithms (geNorm, NormFinder, BestKeeper and the ΔCt method), we found that the rankings of all the reference genes differed among the results of the different algorithms. This variation was expected, however, because the four approaches involve the use of different calculation algorithms, each of which has been verified in many previous studies39,40,41.
In this study, for further verification of the best reference genes, we compared the CVs of the FPKMs of 12 candidate reference genes between the control and different organs in our RNA sequencing (RNA-seq) data (Fig. S2). At the same time, the CVs of the 12 candidates were also compared with our results obtained by geNorm, NormFinder and BestKeeper (Fig. S2), and the overall results were generally consistent. However, this did not undermine the final results of the experiments, because the sequencing data were used only for verification in subsequent validation experiments instead of being directly used to draw conclusions. For this reason, the data could be considered only preliminary, which indicated the trends the FPKM CV values of 12 candidate reference genes. We validated the selected reference genes according to the relative expression of WRKY in the roots of plants under low-temperature stress and that of WRKY in the roots of plants under salt stress. The results shown in Fig. 3 confirm that the candidate reference genes were applicable and stably expressed in Caucasian clover. The results also indicated that the most stable reference genes differed among organs and among treatments, even within the same organ.
Conclusion
Twelve candidate reference genes were selected for qRT-PCR standardization evaluation, and five different statistical methods were used. The results showed that RCD1, PPIL3, NLI1 and NLI2 were the most stable reference genes, while bZIP and EFTu-GTP were the most unstable reference genes. The stable reference genes identified in this report will enhance the accuracy of qRT-qPCR-based analysis of target gene expression and can be used to study related functional genes in Caucasian clover.
References
Heaffer, C. C. & Marten, G. C. Kura clover forage yield, forage quality, and stand dynamics. Can. J. Plant Sci. 71, 1169–1172 (1991).
Alexander, J. R., Baker, J. M., Venterea, R. T. & Coulter, J. A. Kura clover living mulch reduces fertilizer n requirements and increases profitability of maize. Agron Basel 9, 432–447 (2019).
Black, A. D. & Lucas, R. J. In Caucasian clover was more productive than white clover in grass mixtures under drought conditions. in Proceedings of the New Zealand Grassland Association (2000).
Ren, R. et al. Selection and validation of suitable reference genes for RT-qPCR analysis in dove tree (Davidia involucrataBaill.). Trees 33, 837–849 (2019).
Saddhe, A. A., Malvankar, M. R. & Kumar, K. Selection of reference genes for quantitative real-time PCR analysis in halophytic plant Rhizophora apiculata. Peerj 6, e5226 (2018).
Sun, H., Jiang, X., Sun, M., Cong, H. & Qiao, F. Evaluation of reference genes for normalizing RT-qPCR in leaves and suspension cells of Cephalotaxus hainanensis under various stimuli. Plant Methods 15, 31 (2019).
Wang, Z. L. et al. Selection and validation of appropriate reference genes for real-time quantitative PCR analysis in Momordica charantia. Phytochemistry 164, 1–11 (2019).
Lee, J. M., Roche, J. R., Donaghy, D. J., Thrush, A. & Sathish, P. Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). Bmc Mol. Biol. 11, 8 (2010).
Kong, Q. S. et al. Assessment of suitable reference genes for quantitative gene expression studies in melon fruits. Front. Plant Sci. 7, e70603 (2016).
Kong, Q. et al. Evaluation of appropriate reference genes for gene expression normalization during watermelon fruit development. Plos One 10, e0130865 (2015).
Miao, L. et al. Selection of reference genes for quantitative real-time PCR analysis in cucumber (Cucumis sativus L.), pumpkin (Cucurbita moschata Duch.) and cucumber-pumpkin grafted plants. Peerj 7, e6536 (2019).
Wan, H. et al. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 399, 257–261 (2010).
Garcia-Lavina, C. X., Castro-Sowinski, S. & Ramon, A. Reference genes for real-time RT-PCR expression studies in an Antarctic Pseudomonas exposed to different temperature conditions. Extremophiles 23, 625–633 (2019).
Cheng, T. et al. Selection of suitable reference genes for quantitative real-time PCR normalization in Miscanthus lutarioriparia. Mol. Biol. Rep. 46, 4545–4553 (2019).
Lu, J. et al. Selection and validation of reference genes for RT-qPCR analysis of the Ladybird Beetle Henosepilachna vigintioctopunctata. Front. Physiol. 9, 1–11 (2018).
Nong, Q. D. et al. RNA-seq-based selection of reference genes for RT-qPCR analysis of pitaya. FEBS Open Bio. 9, 1403–1412 (2019).
Lin, Y., Zhang, A., Yang, S. & Huang, L. Reference gene selection for real-time quantitative PCR normalization in Hemarthria compressa and Hemarthria altissima leaf tissue. Mol. Biol. Rep. 46, 4763–4769 (2019).
Rafael, N., Ulrik, J., John, M. & German, S. Selection of optimal reference genes for quantitative RT-PCR transcript abundance analysis in white clover (Trifolium repens L.). Funct. Plant Biol. 45, 737 (2018).
Afsana, I., Chris, F. M., Susanna, L., Paul, P. D. & Michael, T. M. Transcription of biotic stress associated genes in white clover (Trifolium repens L.) differs in response to cyst and root-knot nematode infection. Plos One. 10, e0137981 (2016).
Li, B. Z., Han, S. & Zhao, X. RCD1, Oxidative stress regulator, affects the tesponse of Arabidopsis thalianato HgCl2. J. Henan Univ. (Natural ence) 4, 390–393 (2011).
Lidiane, L. B. A. et al. Cowpea and abiotic stresses: Identifcation of reference genes for transcriptional profling by qPCR. Plant Methods. 14, 88 (2018).
Anderson, J. V. et al. Expression of Flowering Locus C and a frame shift mutation of this gene on chromosome 20 differentiate a summer- and winter-annual biotype of Camelina sativa. Plant Direct. 1, 1–14 (2018).
Dekkers, B. J. W. et al. Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 53, 28–37 (2012).
Huggett, J., Dheda, K., Bustin, S. & Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immunity 6, 279–284 (2005).
Guenin, S. et al. Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot 60, 487–493 (2009).
Karuppaiya, P. et al. Identification and validation of superior reference gene for gene expression normalization via RT-qPCR in staminate and pistillate flowers of Jatropha curcas—A biodiesel plant. PLoS ONE 12(5), e0177039 (2017).
Vandesompele, S., et al. Expressed repeat elements improve RT-qPCR normalization across a wide range of zebrafish gene expression studies. PloS ONE 9, (2104).
Liu, D. et al. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS ONE 7, e46451 (2012).
Jia, D. H. et al. Validation of reference genes for quantitative gene expression analysis in Auricularia cornea. J. Microbiol. Methods 163, 7 (2019).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△Ct method. Methods 25, 402–408 (2001).
Wei, W. et al. The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: Identification and expression analysis under biotic and abiotic stresses. Plant Physiol. Biochem. 105, 129–144 (2016).
Wu, K. L., Guo, Z. J., Wang, H. H. & Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26 (2005).
Mangelsen, E. et al. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. Bmc Genomics 9, 194 (2008).
Ling, J. et al. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 12, 471 (2011).
Ayadi, M. et al. The WRKY transcription factor family in citrus: Valuable and useful candidate genes for citrus breeding. Appl. Biochem. Biotech. 180, 516–543 (2016).
Ma, L. et al. Selection and validation of reference genes for quantitative real-time PCR analysis in Iris bulleyana during flower color variation. J. Nucl. Med. 33, 1707–1716 (2019).
Xiao, Z. et al. Selection of reliable reference genes for gene expression studies on Rhododendronmolle G. Don. Front. Plant Sci. 7, 1547 (2016).
Li, W. et al. Selection and validation of appropriate reference genes for quantitative real-time PCR normalization in staminate and perfect flowers of Andromonoecious Taihangiarupestris. Front. Plant Sci. 8, 729 (2017).
Teotia, S., Lamb, R.S. RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana. J. Exp. Bot. 3, 1271–1284 (2011).
Huang, Y. et al. Stable internal reference genes for normalizing real-time quantitative PCR in Baphicacanthus cusia under hormonal stimuli and UV irradiation, and in different plant organs. Front. Plant Sci. 8, 668 (2017).
Tang, W. et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13, 124–131 (2011).
Zhu, Z. & Liang, F. Plant MAP65. Plant Physiol. Commun. 45, 619–624 (2009).
Chen, G. et al. Selection and validation of reference genes for quantitative RT-PCR analysis in Castanea mollissima. J. Plant Physiol. 55, 378–386 (2019).
Udvardi, M. K., Czechowski, T. & Scheible, C. W. R. Eleven golden rules of quantitative RT-PCR. Plant Cell 20, 1736–1737 (2008).
Mascia, T., Santovito, E., Gallitelli, D. & Cillo, F. Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 11, 805–816 (2010).
Funding
This work was supported by the National Natural Science Foundation of China (31802120), the Research and Demonstration of Large-scale Artificial Grassland Combined Plant and Circular Mode (2017YFD0502106), and the Academic Backbone Fund Project of Northeast Agricultural University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yin, X., He, T., Yi, K. et al. Comprehensive evaluation of candidate reference genes for quantitative real-time PCR-based analysis in Caucasian clover. Sci Rep 11, 3269 (2021). https://doi.org/10.1038/s41598-021-82633-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82633-2
- Springer Nature Limited
This article is cited by
-
Transcriptomic and physiological analyses of Trichoderma citrinoviride HT-1 assisted phytoremediation of Cd contaminated water by Phragmites australis
BMC Microbiology (2024)
-
Selection of suitable candidate genes for mRNA expression normalization in bulbil development of Pinellia ternata
Scientific Reports (2022)