Abstract
The complete mitochondrial genome of Lyrognathus crotalus is sequenced, annotated and compared with other spider mitogenomes. It is 13,865 bp long and featured by 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs), 13 protein-coding genes (PCGs), and a control region (CR). Most of the PCGs used ATN start codon except cox3, and nad4 with TTG. Comparative studies indicated the use of TTG, TTA, TTT, GTG, CTG, CTA as start codons by few PCGs. Most of the tRNAs were truncated and do not fold into the typical cloverleaf structure. Further, the motif (CATATA) was detected in CR of nine species including L. crotalus. The gene arrangement of L. crotalus compared with ancestral arthropod showed the transposition of five tRNAs and one tandem duplication random loss (TDRL) event. Five plesiomophic gene blocks (A-E) were identified, of which, four (A, B, D, E) retained in all taxa except family Salticidae. However, block C was retained in Mygalomorphae and two families of Araneomorphae (Hypochilidae and Pholcidae). Out of 146 derived gene boundaries in all taxa, 15 synapomorphic gene boundaries were identified. TreeREx analysis also revealed the transposition of trnI, which makes three derived boundaries and congruent with the result of the gene boundary mapping. Maximum likelihood and Bayesian inference showed similar topologies and congruent with morphology, and previously reported multi-gene phylogeny. However, the Gene-Order based phylogeny showed sister relationship of L. crotalus with two Araneomorphae family members (Hypochilidae and Pholcidae) and other Mygalomorphae species.
Similar content being viewed by others
Introduction
The order Araneae is classified into two infra-orders Mesothelae (primitive spiders) and Opisthothelae (modern spiders). The infra-order Opisthothelae is further classified into two suborders Mygalomorphae and Araneomorphae with 117 families. The family Theraphosidae belongs to suborder Mygalomorphae of infra-order Opisthothelae with 992 species. Out of 992 species, 52 species are known from India1. The members of family Theraphosidae are commonly known as tarantulas or giant spiders for their huge body size. These giant spiders play an important role in controlling the insects2 and also predators on vertebrates and invertebrates3. The venom of these tarantulas is the main source of pharmacological research3.
The Pet trade of tarantulas across the globe is in great demand due to their body size, attractiveness, longitivity and for economic point of view4. So far, 13 species in three genera (Poecilotheria, Thrigmopoeus and Lyrognathus) from India have been reported in the pet trade4. The species Lyrognathus crotalus is endemic to India restricted to northeast region and frequently traded. The correct identification of these tarantulas is the basic need due to their economical and medicinal values and involvement in the pet trade. However, identification of species in the absence of well-preserved specimens is not possible through morphology alone. Nowadays, molecular data are widely employed for identification of the species, resolving taxonomic ambiguities, and to infer the phylogenetic relationships. The multi-gene based analysis revealed the monophyly of Mesothelae, Opisthothelae, Mygalomorphae and Araneomorphae with adequate support and recovered Hypochilidae and Filistatidae as sister groups5,6. However, the phylogeny using mitogenome data, including different tree building methods, and gene orders has never been attempted. The mitochondrial genome is featured by their maternal legacy, high level of evolution and low rate of intermolecular genetic recombination, which is widely used in phylogenetic studies, population genetics, and phylogeography7. The circular mitochondrial genome of arthropods usually 14–19 kb in size, with 37 genes (22 transfer RNA genes, two ribosomal RNA genes, 13 protein-coding genes), and a noncoding control region. Till now, 32 mitochondrial genomes of 15 spider families are accessible in the GenBank database (https://www.ncbi.nlm.nih.gov/)8,9,10,11,12,13,14,15,16,17,18. In the present study, we sequenced the complete mitochondrial genome of L. crotalus using Next-generation sequencing. This is the second mitochondrial genome sequenced in the family Theraphosidae. We compared this denovo mitochondrial genome with 16 spider species mitochondrial genomes in 15 families to observe the codon usage patterns, gene features, gene arrangements, secondary structure of tRNAs and control region (CR), and their phylogenetic relationships.
Materials and Methods
Sample collection, and DNA extraction
The specimen of Lyrognathus crotalus was collected from the Badarpur (24.85N 92.56E), Assam, Northeast India. The morphological identification of this specimen was done by published taxonomic keys19, and stored in absolute ethyl alcohol at −80 °C in Centre for DNA Taxonomy, Molecular Systematics Division, Zoological Survey of India, Kolkata. DNeasy DNA Extraction kit (Qiagen, Valencia, CA) was used for genomic DNA extraction and quantified by dsDNA high-sensitivity kit (Thermo Fisher Scientific, MA, USA) in Qubit fluorometer. In this study, no prior permission was required for the collection as the species is neither endangered nor protected species in IUCN Red List or Indian Wildlife Protection Act, 1972.
Mitochondrial genome sequencing and assembly
The Genotypic Technology Pvt. Ltd. Bangalore, India (http://www.genotypic.co.in/) had carried out the sequencing. The sequencing and assembly protocol was followed by our previous study20,21. The whole genome library of genomic DNA was sequenced using Illumina Hiseq. 2500 (2 × 150 base paired-end reads) (Illumina, USA) platform which yielded ~14 million reads. The TruSeq DNA Library Preparation kit (https://support.illumina.com/downloads/truseq) was used for the construction of the paired-end library with standard protocols. The trimming and filtering of the raw sequencing reads were done by using the NGS-Toolkit22 to removing adapter contamination and low-quality reads with a cutoff of Phred quality scores of Q20. Burrows-Wheeler Alignment (BWA) tool23 screened the high quality reads (1.4 million) using Seqtk (https://github.com/lh3/seqtk) and down sampled high-quality reads and assembled with SPAdes 3.9.024, using Ornithoctonus huwena mitochondrial genome (NC_005925.1) as a reference.
Annotation, secondary structure prediction, and comparative studies
The annotation of PCGs and rRNAs were done by using MITOS web-server25. The annotation of the tRNA genes was the most difficult steps to define their location and gene boundaries. To search the tRNAs in the mitochondrial genome, the tRNAscan-SE 1.2126 was used, but incapable to detect tRNAs. Further, we have used other spider tRNA sequences from GenBank to confirm the locations and boundaries of each tRNAs. Further, the anticodon arm motifs (or sometimes only the 3-bp anticodon sequence) were examined manually which were conserved among all spiders.
The start and stop codons of PCGs were observed by using the ClustalX program27. The PCGs of L. crotalus with other spider species were aligned by MEGAX28. For acquiring the accession number of the complete annotated L. crotalus mitochondrial genome from GenBank, the Sequin submission tool (http://www.ncbi.nlm.nih.gov/Sequin/) was used. The circular image of L. crotalus mitochondrial genome was drawn by using online server CGView29 (http://stothard.afns.ualberta.ca/cgview_server/). The length and locations of spacer regions (overlapping and intergenic) of L. crotalus mitochondrial genome were detected manually.
The nucleotide composition, codon usages, relative synonymous codon usage (RSCU) was done by MEGAX. To calculate the skewness, we used the formula: AT skew = (A − T)/(A + T) and GC skew = (G − C)/(G + C)30. Codon usage bias was evaluated by calculating of effective number of codon (ENc) with the DnaSP6.031. The relative synonymous codon usage (RSCU) graph is plotted in Microsoft Office Excel. The ratios of non-synonymous substitutions (Ka) and synonymous (Ks) substitutions were estimated in DnaSP6.0. The transition and transversion ratio versus genetic distance calculated by using DAMBE532. We predicted the secondary structure of tRNAs and CR for L. crotalus mitochondrial genome. The tRNAs secondary structures were predicted by VARNA 3.9333. The prediction of secondary structure of CR was done by The Mfold web server34.
Preparation of data sets, model selection, phylogenetic analyses
Out of 32 available spider mitogenomes in the global database, 16 species mitochondrial genomes were retrieved and used in the present dataset based on representative families8,9,10,11,12,13,14,15,16,17,18. The mitogenome of Limulus polyphemus (order: Xiphosura) were also retrieved from GenBank and used as an out-group35 (Table S1). The four data sets were prepared for phylogenetic analysis: (1) PCGs without GBlock36, 11453 bp; (2) PCGs without GBlock (third codon position excluded), 7640 bp; (3) PCGs with GBlock, 8706 bp; (4) PCGs with GBlock (third codon position excluded), 5804 bp. The PartitionFinder version 2.1.137, with the greedy algorithm was used to find the best substitution models for Bayesian Inference (BI) and maximum likelihood (ML). The PartitionFinder analyses: codon positions for each PCGs (13 genes × 3 codons = 39 partitions), PCGs excluding 3rd codon position (13 genes × 2 codons = 26 partitions) (Table S2). We used the online web portal The CIPRES Science Gateway v.3.1 (www.phylo.org/sub_sections/portal/) to perform Mr. Bayes 3.2 for BI analysis38 and ML analysis using IQ-tree web server using four data sets39. The phylogenetic tree was visualized and edited using FigTree v1. 4.240 (http://tree.bio.ed.ac.uk/software/figtree/). We used the MLGO web server41 for gene order based phylogeny (http://www.geneorder.org/server.php) (Table S3).
Gene arrangement analysis
Three methods were applied to see the gene arrangement scenario: (1) CREx42 based on the common interval; (2) mapping the gene boundaries on the gene order to see the unique, derived and ancestral gene boundaries between the ancestral arthropod L. polyphemus and other taxa of spiders (Tables S4 and S5); and (3) TreeREx Analysis43 based on common used to evaluate evolutionary pathways. TreeREx analysis is the extended version of CREx with default settings was performed: strong consistency method applied (-s); weak consistency method applied (-w); parsimonious weak consistency method applied (-W); get alternative bp scenario for prime nodes (-o); maximum number of inversions (-m = 0) + TDRL scenarios considered. Three levels of nodes were inferred, green colour nodes were consistent, yellow colour 1-consistent nodes, and the red colour fallback nodes.
Results and Discussion
Genome structure, organization and composition
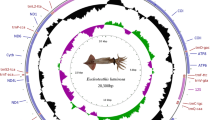
L. crotalus complete mitochondrial genome (accession number MN072398) is 13,866 base pairs (bp) in length. This is the smallest mitochondrial genome of spider among all published mitochondrial genomes so far due to the presence of extremely truncated tRNAs. It included 37 genes: 13 PCGs, large and small rRNAs, 22 tRNAs and one non-coding region (D-Loop) with the origin of light-strand replication (OL) (Fig. 1, Table 1). The majority strand contains 22 genes and minority with 15 genes (Table 1). The AT and GC content of nucleotide was 68.74% and 31.26%, respectively (Table S6), like other spider mitochondrial genomes. The highest AT content observed in tRNAs (69.68%,), followed by PCGs (68.72%), rRNAs (68.66%), and CR (67.67%). The mitochondrial genome showed positive AT (0.07) and negative GC (-0.36) skewness (Table S6) in contrast to other spider mitochondrial genome.
The circular representation of the complete mitochondrial genome of L. crotalus. Direction of gene transcription is indicated by arrows in entire complete genome. PCGs are shown as purple arrows, rRNA genes as pink arrows, tRNA genes as peach color arrows and CR regions as gray rectangles. The GC content is plotted using a black sliding window, as the deviation from the average GC content of the entire sequence. GC-skew is plotted using a colored sliding window (green and orchid color), as the deviation from the average GC skew of the entire sequence. The figure was drawn using CGView online server (http://stothard.afns.ualberta.ca/cgview_server/) with default parameters. The species photograph was taken by the fourth author using Leica Microscope EZ4 HD with Leica software application suite (LAS EZ) and edited manually in Adobe Photoshop CS 8.0.
Protein-coding genes
The total length of PCGs was 10,587 bp in L. crotalus. The ATN start codons were used by most of the PCGs except cox3, and nad4 with TTG. Comparative study revealed that ATN and TTG start codon used by most of the PCGs of spider mitochondrial genomes. TTT start codon used by only cox1 (Tetragnatha maxillosa17); TTA used by cox1 (Phyxioschema suthepium, Ornithoctonus huwena8, Agelena silvatica11, Carrhotus xanthogramma13, Selenops bursarius16, Oxytate striatipes15); GTG used by cox2 (Selenops bursarius, Pholcus phalangioides), cytb (Ornithoctonus huwena), nad2 (Pholcus phalangioides), nad6 (Oxyopes sertatus14), atp6 (Pholcus phalangioides); CTG used by cox1 (Neoscona theisi10, Liphistius erawan); CTA used by cox1 (Calisoga longitarsis8). The stop codons TAA, TAG, and T(AA) were used commonly by most of the PCGs as observed in other spiders (Table S7).
Codon usage bias and mutations
The use of codons or codon usage bias is a fundamental phenomenon in nature44,45. The main influencing forces for codon usages are the mutation pressure and natural selection. Codon usage bias can be triggered by a number of other factors such as the content of nucleotides, gene length and their function, and the external environment45. We investigated the GC content to study the nucleotide distribution of all three codon positions of PCGs for 17 spider mitochondrial genomes (Fig. S1). The average GC content was 29.02%, while the value of GC1s, GC2s and GC3s were 35.98%, 34.90% and 20.02% respectively. The codon frequency ending with A/T is higher than G/C due to the AT rich segments which leads to the high codon bias46,47. The comparative study of spider mitochondrial genomes revealed 21 codons (9 with A-ending, 12 with U-, and none with G- or C-ending) with high frequency (Table S8). This result suggested that compositional constrain may play an important role in the codon usage patterns in spider species.
The average of the Effective Codon number (ENc) values for all the PCGs was 42.97, which indicate a strong codon bias, ranging from 39.31 to 46.83. We plotted ENc-GC3s value to explain the relationship between nucleotide composition and codon bias (Figs. 2A and S2). The result specified that not only mutation, but other factors like natural selection might be involved in shaping the codon bias in spider mitochondrial genomes. To confirm the relation between GC12 and GC3 and explain the mutation-selection equilibrium in shaping the codon usage bias, we used neutrality plot analysis (Fig. 2B). This plot indicates that the genes have a wide range of GC3 value distributions, ranging from 8.1% to 34.7%, and also showed the positive correlation between GC12 and GC3 (r = 8455, p < 0.01). Moreover, the slope of the regression line of the entire coding sequences was 0.2516. So, the natural selection may probably dominate the codon bias rather than mutation bias48.
(A) ENC versus GC3 plotting of concatenated PCGs of spider mitochondrial genomes; (B) Neutrality plot of concatenated PCGs of spider mitochondrial genomes; (C) Box plot for pairwise divergence of Ka/Ks ratio for 13 PCGs of spider mitochondrial genomes; (D) Genetic distance versus transition-transversion ratio of 13 PCGs spider mitochondrial genomes. The figure was edited in Adobe Photoshop CS 8.0.
Non-synonymous and synonymous substitutions
To investigate the selective pressure and evolutionary relation of the homogenous or heterogeneous species, non-synonymous and synonymous substitutions (Ka/Ks) ratio was used49. Our result showed the average Ka/Ks ratio ranging from 0.122 ± 0.03 in cox1 to 0.443 ± 0.14 in nad6 gene and the resulted following order: cox1 < nad1 < cox2 < nad5 < cytb < cox3 < nad4 < atp6 < nad4l < nad3 < nad2 < nad6 < atp8. This result indicated that the 13 PCGs excluding atp8 of all spider mitochondrial genomes including L. crotalus were evolving under purifying selection (Fig. 2C). The value of Ka/Ks for all the PCGs was below one, indicating the mutations swapped by synonymous substitutions. The cox1 gene with low Ka/Ks ratio represent fewer changes in amino acids and hence widely used as a potential molecular marker for species identification and phylogenetic analysis50,51. The transversion and transition plot against the genetic distance showed a linear relationship for PCGs (Fig. 2D). The value of the substitution saturation index for the combined dataset of all PCGs of 17 spider mitochondrial genomes (Iss = 0.4261) was significantly lower than the critical values (Iss. cSym = 0.8367 or Iss.cAsym = 0.6659). Hence, the combined data is suitable for phylogenetic analysis.
Ribosomal and transfer RNA genes
Two rRNAs were observed in L. crotalus and other spider mitochondrial genome. The large ribosomal rrnL (16S RNA) placed between trnV and trnL1, was 1128 bp long; the small rrnS (12S RNA) between trnI and trnV, was 627 bp long (Table 1). L. crotalus has 22 tRNAs (total length 1,329 bp), ranging from 41 to 99 bp in length. (Table S5).
The prediction of the secondary structure and gene boundaries for all the tRNA genes was extremely difficult, as truncated at their 3′ end and lack of proper base pairing at the 5′ end (Fig. S3). The extreme truncation or atypical structure of tRNA is very peculiar in Arachnids mitochondrial genomes, including Araneae9,52. Seventeen of the 22 tRNAs has atypical secondary structures, missing either a D-arm or T-arm. The majority of tRNAs also have mismatches on T-arm, D-arm, acceptor arm or anticodon arm. The lack of acceptor arm or very poorly paired arm was observed in the following genes (trnL1, trnS1, trnE, trnL2, trnI). The lack of T-arm and loop were observed in 13 genes (trnC, trnD, trnF, trnG, trnH, trnK, trnL2, trnN, trnP, trnI, trnY, trnQ and trnV), which was inferred by TV-replacement loops. The lack of DHU loop observed in trnN whereas, lack of DHU arm and loop observed in trnS1, trnI (Fig. S3). Out of 22 tRNAs, boundaries of nine genes were overlapped with the adjacent gene on both the ends. The gene overlaps or quantity of truncation in each tRNA gene were differ from species to species. trnE showed overlaps of 39 nucleotides (nt) with its neighboring gene (trnF) on the same stand at 5′ end and 26 nt overlap with trnR gene on the opposite strand at the 3′ end and. After overlapping on both the ends, only four nt was remaining which exclusively denoted for trnE. trnC showed overlaps of 28 nt at the 5′ end with its neighboring gene (cox1) on the opposite strand and 35 nt overlap at the 3′ end with trnY gene on the same strand. After overlapping on both the ends, no nucleotide was remaining for trnC.
The A + T-rich region
The A + T-rich region in mitochondrial genome is important for the initiation of replication in Metazoans53,54, and also called the control region (CR). It is located between trnM and trnQ in L.crotalus, spans 356 bp with 67.67% AT content and showed positive AT/GC skew (0.04/0.15), indicating an obvious bias towards the use of A and G. The non-coding region named replication origin of L-strand (OL) region was also found. The “OL” region (TCCTCCTCCGCGGAAAAGAGAGGAGGA) is 27 bp in length and has the potential to fold into a stem-loop secondary structure (Fig. 3). Apart from the conserved elements in A + T-rich region, tandem repeats were also found to be a characteristic of A + T-rich regions. In L.crotalus, two tandem repeats of 10 bp (ATTTTTATTC) and six tandem repeats of 6 bp (CATATA) were present at the 3′ end upstream of the trnQ. Further, this motif was also detected in other eight species (A. angulatus, C. longitarsis, C. xanthogramma, H. thorelli, N. theisi, O. huwena, P. phalangioides, S. bursarius). All elements which are related to the regulation of transcription and control of DNA replication in the A + T-rich region were arranged in the conserved order as compared to other Opisthothele spiders.
Phylogenetic analyses
Eight phylogenetic trees based on four datasets and two inference methods, Maximum likelihood (ML) and Bayesian Inference (BI) were generated (Figs. 4 and S4–S6). All the analyses supported the monophyly of suborders Opisthothelae with high bootstrap support and posterior probability. The infraorder Mygalomorphae was recovered as monoplyletic in all the analyses. The Araneomorphae was recovered paraphyletic in all the analyses except BI-2, BI-4, ML-4 (Fig. 4). The paraphyly in the Araneomorphae is due to the varied position of family Hypochilidae and Pholcidae. The family Hypochilidae is first to branch from the other taxa of spiders and leaving two clade with strong posterior probabilities and bootstrap support in BI-1, 3, ML-2 (Figs. S4 and S6). The first clade of Mygalomorphae + family Pholcidae of Araneomorphae and; second clade of Araneomorphae. In two analyses (ML-1, 3), the families Pholcidae and Hypochilidae were grouped with Mygalomorphae taxa with low bootstrap support (Fig. S5). In all the analyses L. crotalus is cladded with O. huwena and showed sister relationship with C. longitarsis (Nemesiidae) and P. suthepium (Dipluridae). The estimated tree from the mitochondrial genome is well supported by previously generated multilocus phylogeny5. However, the genital structure is the distinguishable feature of the Mygalomorphae and Araneomorphae spiders with three types of genitalia5,55: Haplogyne (simple) in Mygalomorphae, Entelegyne (two separate ducts for copulation and fertilization) and non-entelegyne (single duct for both copulation and fertilization) in Araneomorphae. The estimated trees in this study are also supported by a previous study showing the monophyly of entelygyne taxa. However, non-entelygyne Araneomorphae (Pholcidae and Hypochilidae) without cylindrical gland spigots always grouped with Haplogyne (Mygalomorphae).
Linearized view of gene arrangements of spider species and ancestor (Limulus polyphemus) in correspondence with Gene order based tree MLGO phylogeny. Species with identical gene order in correspondence to MLGO phylogeny. The PCGs and rRNAs are represented by their standard nomenclature with light orange and green color respectively. The rRNAs are represented by dark grey color. The CRs are represented by pink color. Derived gene boundaries are numbered with numerals, and plesiomorphic gene block are underline and numbered with alphabet. The figure was edited in Adobe Photoshop CS 8.0.
The gene order based MLGO phylogeny indicated the monophyly of suborder Opisthothelae (Fig. 5, Table S3). The family Salticidae and Agelenidae of Araneomorphae showed sister relationship with other spider taxa. The Further two clades were formed: 1) Mygalomorphae + Hypochilidae and Pholcidae; 2) other Araneomorphae taxa. The close relationship of Araneomorphae non-entelegyne taxa (Pholcidae and Hypochilidae) with Mygalomorphae was also indicated in the sequence based phylogenetic analyses. The studied tarantula species, L. crotalus revealed a close relationship with the Hypochilidae and Pholcidae in the MLGO phylogeny as they share the same gene order. MLGO phylogeny also revealed paraphyly of entelygyne taxa5. The BA and ML phylogenies are discordant with MLGO phylogeny and indicated the cladding of L. crotalus (Theraphosidae) with non-entelegyne taxa. However, more mitochondrial genome data of both non- entelegyne and haplogyne taxa would cover the consistent sign of deep phylogenetic relationship between them.
Gene arrangement
Three methods were applied to see the gene arrangement (1) CREx; (2) mapping the gene boundaries on the gene order; (3) TreeREx Analysis.
CREx
The transpositions, inversions, and inverse transpositions are the characteristic pattern of gene arrangement in the mitochondrial genome56. To explain the transpositions in gene arrangement, the Tandem Duplication–Random Loss (TDRL) is the most commonly known process42. By comparing the gene order of L. crotalus with the putative ancestral arthropod mitochondrial genome (L. polyphemus) by using CREx analysis, five tRNAs (trnT, trnL2, trnI, trnQ, and trnY) were found to transposition and one TDRL event in block trnN-trnA-trnS1-trnR. The trnI changed its position to a new location between rrnS and trnQ and also moved from majority to minority strand. Inverse transposition of trnI was also observed in A. silvatica, C. longitarsis, O. huwena, and P. suthepium. The comparative study of gene arrangement revealed that L. crotalus (Theraphosidae) shared the gene order with following species, O. huwena (Theraphosidae), P. suthepium (Dipluridae), C. longitarsis (Nemisiidae), H. thorelli (Hypochilidae) and P. phalangioides (Pholcidae). Further, CREx analysis revealed the inversion of trnI within mygalomophs, Hypochilidae and Pholcidae. Hence, the studied species L. crotalus is more inclined towards family Hypochilidae and Pholcidae (Table S4).
Mapping of the gene boundaries
The mapping of the gene boundaries of the ancestral gene order revealed the five plesiomorphy boundaries (A-E) which were represented in the Figs. 5 and 6A. The plesiomorphic block A (cox1-cox2-trnK-trnD-atp8-atp6-cox3-trnG-nad3), B (trnF-nad5-trnH-nad4-nad4L), D (rrnL-trnV-rrnS) and E (nad2-trnW) are retained in all taxa of Mygalomorphae and Araneomorphae except family Salticidae (E lost). The block C (trnP-nad6-cytb) is retained in Mygalomorphae and two families of Araneomorphae (Pholcidae and Hypochilidae) (Fig. 6A, Table S5). We have identified 146 derived gene boundaries, including 11 unique boundaries in 16 species. Twenty two derived boundaries were repeated 135 times in 16 species. In spiders, most of the taxa have an identical gene arrangement, though they belong to different families. The gene arrangement of the P. phalangioides (Pholcidae) is exactly identical to H. thorelli (Hypochilidae) and L. crotalus (Theraphosidae) shared 20 boundaries. Further, gene arrangement of other mygalomorphae taxa, C. longitarsis (Nemesiidae), O. huwena (Theraphosidae), and P. suthepium (Dipluridae) are identical and shared 20 boundaries. The gene arrangements of Araneomorhae taxa O. sertatus (Oxyopidae), W. fidelis (Lycosidae), O. striatipes (Thomisidae), T. maxillosa (Tetragnathidae) A. angulatus, N. theisi (Araneidae) are identical and shared 21 boundaries. The derived gene boundaries 1, 7–11, 13, 14, 16, 17 are synapomorphy for Opisthothelae spiders. The gene boundaries 22 and 23 (rrnS-trnI, trnI-trnQ) are synapomorphy for the Mygalomorphae + Pholcidae and Hypochilidae clade. Gene boundaries 5, 6 and 12 (nad6-trnI, trnI-cytb, rrnS-trnQ) are synapomorphy for Araneomorphae clade except families Pholcidae and Hypochilidae.
(A) Mapping genome rearrangements and reductions onto an evolutionary tree of the spiders. Maximum likelihood tree (ML-1) is used for representation of the plesiomorphic and synapomorphic gene blocks. Synapomorphic gene blocks are labelled with numerals, and plesiomorphic with alphabet of different colours. (B) Original output of TreeREx analysis of spiders gene order. Maximum likelihood tree (ML-1) is used for representation and analysis. The rearrangements on the branches are given as T for a transposition and TDRL for tandem-duplication-random-loss events; green nodes marks consistent reconstructed nodes; and red nodes are reconstructed with the fallback method. The value following the “P” in the node label shows how much better the chosen solution is in comparison with other possible solution(s). The figure was edited in Adobe Photoshop CS 8.0.
TreeREx analysis
TreeREx detected 15 nodes, 13 consistent, two inconsistent with four transpositions, seven inversions and one inverse transpositions (Fig. 6B). The transposition of trnI at node A14 towards A4 node makes two new gene boundaries, 22 and 23 and become a synapomorphy character for Mygalomorphae and two families of Araneomorphae (Hypochilidae, Pholcidae). The inversion of trnI on A3 node towards A2 separated the Mygalomorphae from Pholcidae and Hypochilidae and once more inversion of trnI towards L. crotalus separated the L. crotalus to O. huwena. Three gene boundaries (5, 6, 12) are synapomorphy for Araneomorphae clade. The inversion of trnN on node A10 towards A5 separated the families Dictynidae and Agelenidae from Thomisidae, Lycosidae, Oxyopidae, Selenopidae, and Salticidae. The inverse transposition of trnN towards Dictynidae and transposition of (trnL2, trnA, trnN) and inversion of trnI towards Agelenidae separated these two families from each other. The inversions of trnL2 and trnC at node A6 towards Selenopidae and transposition of trnE and trnW towards Salticidae separated these two families from each other.
Conclusion
The complete mitochondrial genome of endemic giant tarantula L. crotalus was characterized and compared with other members of the Araneae. The phylogenetic relationships based on mitochondrial genome data are congruent with morphology and the earlier multiple marker gene results. The Araneomorphae and Mygalomorphae are recovered as monophyletic as previously reported. On the other hand, based on morphology, tarantulas (family Theraphosidae) are more inclined to plagiognath condition (chelicerae are intermediate position, neither parallel nor opposing each other). The gene order based analysis of L. crotalus also indicated the close relationship with Araneomorphae (Hypochilidae and Pholcidae) and other Mygalomorphae species. However, in-depth data on more taxa of Araneae could build up our knowledge towards the rearrangement and evolutionary events.
References
World Spider Catalog. World Spider Catalog. Version 20.0. Natural History Museum Bern. Avialable online at, http://wsc.nmbe.ch, (Accessed on 1st December 2018).
Marc, P., Canard, A. & Ysnel, F. Spiders (Araneae) useful for pest limitation and bioindication. Agric. Ecosyst. Environ. 74, 229–273 (1999).
Escoubas, P. & Rash, L. Tarantulas: eight-legged pharmacists and combinatorial chemists. Toxicon. 43, 555–574 (2004).
Siliwal, M., Molur, S. & Raven, R. Mygalomorphs of India: An overview. ENVIS Bulletin: Arthropods and their Conservation in India (Insects & Spiders) 14, 1 (2013).
Wheeler, W. C. et al. The spider tree of life: phylogeny of Araneae based on target-gene analyses from an extensive taxon sampling. Cladistics. 33, 574–616 (2016).
Garrison, N. L. et al. Spider phylogenomics: untangling the spider tree of life. Peer J. 4, e1719 (2016).
Simon, C., Buckley, T. R., Frati, F., Stewart, J. B. & Beckenbach, A. T. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 37, 545–579 (2006).
Qiu, Y., Song, D., Zhou, K. & Sun, H. The Mitochondrial Sequences of Heptathela hangzhouensis and Ornithoctonus huwena Reveal Unique Gene Arrangements and Atypical tRNAs. J. Mol. Evol. 60, 57–71 (2005).
Masta, S. E. & Boore, J. L. Parallel evolution of truncated transfer RNA genes in arachnid mitochondrial genomes. Mol. Biol. Evol. 25, 949–959 (2008).
Li, C., Wang, Z. L., Fang, W. Y. & Yu, X. P. The complete mitochondrial genome of the orb-weaving spider Neoscona theisi (Walckenaer) (Araneae: Araneidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 4035–4036 (2016).
Zhu, X. L. & Zhang, Z. S. The complete mitochondrial genome of Agelena silvatica (Araneae: Agelenidae). Mitochondrial DNA Part B. 2, 58–59 (2017).
Liu, M., Zhang, Z. & Peng, Z. The mitochondrial genome of the water spider Argyroneta aquatica (Araneae: Cybaeidae). Zool. Scr. 44, 179–190 (2014).
Fang, W. Y., Wang, Z. L., Li, C., Yang, X. Q. & Yu, X. P. The complete mitogenome of a jumping spider Carrhotus xanthogramma (Araneae: Salticidae) and comparative analysis in four salticid mitogenomes. Genetica. 144, 699–709 (2016).
Pan, W. J., Fang, H. Y., Zhang, P. & Pan, H. C. The complete mitochondrial genome of striped lynx spider Oxyopes sertatus (Araneae: Oxyopidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 1616–1617 (2016).
Kim, J. Y., Yoo, J. S. & Park, Y. C. The complete mitochondrial genome of the green crab spider Oxytate striatipes (Araneae: Thomisidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 1878–1879 (2016).
Pan, W. J., Fang, H. Y., Zhang, P. & Pan, H. C. The complete mitochondrial genome of flat spider Selenops bursarius (Araneae: Selenopidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 1488–1489 (2016).
Wang, Z. L., Li, C., Fang, W. Y. & Yu, X. P. The Complete Mitochondrial Genome of two Tetragnatha Spiders (Araneae: Tetragnathidae): Severe Truncation of tRNAs and Novel Gene Rearrangements in Araneae. Int. J. Biol. Sci. 12, 109–121 (2016).
Wang, Z. L., Li, C., Fang, W. Y. & Yu, X. P. The complete mitochondrial genome of the wolf spider Wadicosa fidelis (Araneae: Lycosidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 3909–3910 (2016).
West, R. C., Nunn, S. C. & Hogg, S. A new tarantula genus, Psednocnemis, from west Malaysia (Araneae: Theraphosidae), with cladistics analysis and biogeography of Selenocosmiinae Simon. Zootaxa. 3299, 1–43 (1889).
Chakraborty, R. et al. The complete mitochondrial genome of Melon thrips, Thrips palmi (Thripinae): Comparative analysis. PLoS One. 13, e0199404 (2018).
Kumar, V. et al. The first complete mitochondrial genome of marigold pest thrips, Neohydatothrips samayunkur (Sericothripinae) and comparative analysis. Sci. Rep. 9, 191 (2019).
Patel, R. K. & Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLos One. 7, e30619 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–1760 (2009).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Bernt, M. et al. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319 (2013).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic. Acids. Res. 25, 955–964 (1997).
Thompson, J. D., Gibson, T. J. & Higgins, D. G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2.3.1–2.3.22 (2002).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Grant, J. R. & Stothard, P. The CGViewServer: a comparative genomics tool for circular genomes. Nucleic. Acids. Res. 36, W181–W184 (2008).
Perna, N. T. & Kocher, T. D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41, 353–358 (1995).
Rozas, J. & Rozas, R. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput. Appl. Biosci. 11, 621–625 (1995).
Xia, X. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 30, 1720–1728 (2013).
Darty, K., Denise, A. & Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 25, 1974–1975 (2009).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic. Acids. Res. 31, 3406–3415 (2003).
Lavrov, D. V., Boore, J. L. & Brown, W. M. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol. Biol. Evol. 17, 813–824 (2000).
Abascal, F., Zardoya, R. & Telford, M. J. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic. Acids. Res. 38, W7–W13 (2010).
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T. & Calcott, B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773 (2017).
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees in Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA. pp 1–8 (14 Nov. 2010).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Rambaut, A. FigTree. Version 1.4.2, Inst. Evol. Biol., Univ. Edinburgh (2014).
Hu, F., Lin, Y. & Tang, J. MLGO: phylogeny reconstruction and ancestral inference from gene-order data. BMC Bioinformatics. 15, 354 (2014).
Bernt, M. et al. CREx: Inferring Genomic Rearrangements Based on Common Intervals. Bioinformatics. 23, 2957–2958 (2007).
Bernt, M., Merkle, D. & Middendorf, M. An Algorithm for Inferring Mitochondrial Genome Rearrangements in a Phylogenetic Tree. Comparative Genomics International Workshop, RECOMB-CG 2008, 5267 Lecture Notes in Bioinformatics (LNBI), 143–157 (2008).
Chakraborty, S., Uddin, A. & Choudhury, M. N. Factors affecting the codon usage bias of SRY gene acrossmammals. Gene. 630, 13–20 (2017).
Whittle, C. A. & Extavour, C. G. Expression-Linked Patterns of Codon Usage, Amino Acid Frequency, and Protein Length in the Basally Branching Arthropod Parasteatoda tepidariorum. Genome Biol. Evol. 8, 2722–2736 (2016).
Mondal, S. K., Kundu, S., Das, R. & Roy, S. J. Analysis of phylogeny and codon usage bias and relationship of GC content, amino acid composition with expression of the structural nif genes. Biomol. Struct. Dyn. 34, 1649–66 (2016).
Wei, L. et al. Analysis of codon usage bias of mitochondrial genome in Bombyx mori and its relation to evolution. BMC Evol. Biol. 14, 262 (2014).
Sueoka, N. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. USA 85, 2653–2657 (1988).
Hu, T. & Banzhaf, W. Nonsynonymous to Synonymous Substitution Ratio ka/ks: Measurement for Rate of Evolution in Evolutionary Computation. Springer-Verlag Berlin Heidelberg. 5199, 448–457 (2008).
Astrin, J. J. et al. Towards a DNA Barcode Reference Database for Spiders and Harvestmen of Germany. PLoS One. 11, e0162624 (2016).
Barrett, R. D. H. & Hebert, P. D. N. Identifying spiders through DNA barcodes. Can. J. Zool. 83, 481–491 (2005).
Masta, S. E. & Boore, J. L. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol. Biol. Evol. 21, 893–902 (2004).
Zhang, D. X. & Hewitt, G. M. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary Studies. Biochem. Syst. Ecol. 25, 99–120 (1997).
Saito, S., Tamura, K. & Aotsuka, T. Replication origin of mitochondrial DNA in insects. Genetics. 171, 1695–1705 (2005).
Ramirez, M. J. The Morphology and phylogeny of dionychan spiders (Araneae: Araneomorphae). Bull. Am. Mus. Nat. Hist. 390, 1–374 (2014).
Cameron, S. L. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 59, 95–117 (2014).
Acknowledgements
The authors are thankful to the Director, Zoological Survey of India, Kolkata, for providing necessary facilities, constant support and encouragement throughout the study. Authors thankful to Mrs. Barnita Kundu for the specimen collection. The study is financially supported by Zoological Survey of India, Kolkata, Ministry of Environment Forest and Climate Change under National Faunal Genome Resources (NFGR) Program.
Author information
Authors and Affiliations
Contributions
K.T., V.K. and R.C. conceived and designed the experiment, P.P. performed taxonomic identification of the spider species and captured photographs, V.K. and K.C. contributed chemicals, K.T. and V.K. generated DNA data, V.K., K.T. R.C. S.K. and I.T. analysed the data, V.K., K.T. and I.T. wrote the manuscript text, and prepared the figures, all authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kumar, V., Tyagi, K., Chakraborty, R. et al. The Complete Mitochondrial Genome of endemic giant tarantula, Lyrognathus crotalus (Araneae: Theraphosidae) and comparative analysis. Sci Rep 10, 74 (2020). https://doi.org/10.1038/s41598-019-57065-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-57065-8
- Springer Nature Limited
This article is cited by
-
Comparative Analysis of the Complete Mitochondrial Genomes of Three Sisoridae (Osteichthyes, Siluriformes) and the Phylogenetic Relationships of Sisoridae
Biochemical Genetics (2024)
-
The complete mitochondrial genome of the intertidal spider (Desis jiaxiangi) provides novel insights into the adaptive evolution of the mitogenome and the evolution of spiders
BMC Ecology and Evolution (2021)
-
Characterization of four mitochondrial genomes of family Neritidae (Gastropoda: Neritimorpha) and insight into its phylogenetic relationships
Scientific Reports (2021)
-
Comparative analysis of the complete mitochondrial genomes in two limpets from Lottiidae (Gastropoda: Patellogastropoda): rare irregular gene rearrangement within Gastropoda
Scientific Reports (2020)