Abstract
Heterosis has been utilized in commercial production, but the heterosis mechanism has remained vague. Hybrid cotton is suitable to dissect the heterosis mechanism. In order to explore the genetic basis of heterosis in Upland cotton, we generated paternal and maternal backcross (BC/P and BC/M) populations. Data for yield and yield-component traits were collected over 2 years in three replicated BC/P field trials and four replicated BC/M field trials. At single-locus level, 26 and 27 QTLs were identified in BC/P and BC/M populations, respectively. Six QTLs shared in both BC populations. A total of 27 heterotic loci were detected. Partial dominant and over-dominant QTLs mainly determined yield heterosis in the BC/P and BC/M populations. QTLs for different traits displayed varied genetic effects in two BC populations. Eleven heterotic loci overlapped with QTLs but no common heterotic locus was detected in both BC populations. We resolved the 333 kb (48 genes) and 516 kb (25 genes) physical intervals based on 16 QTL clusters and 35 common QTLs, respectively, in more than one environment or population. We also identified 189 epistatic QTLs and a number of QTL × environment interactions in two BC populations and the corresponding MPH datasets. The results indicated that cumulative effects contributed to yield heterosis in Upland cotton, including epistasis, QTL × environment interaction, additive, partial dominance and over-dominance.
Similar content being viewed by others
Introduction
Heterosis refers to the phenomenon of F1 hybrids performing better over their parents in yield, quality and adaptation. Dominance, over-dominance and epistasis hypotheses have been proposed to explain the heterosis mechanism. The three hypotheses demonstrated complementarity between dominant alleles and deleterious recessive alleles1,2, superiority of heterozygote3,4 or mimicry over-dominance with repulsion-phase linkage of favorable alleles5,6 and interactions among non-allelic genes7,8,9, respectively. Some previous studies reported the major role of dominance effect on heterosis in rice10 and maize11. However, over-dominance had also been detected as the primary genetic basis of heterosis for decades, such as in maize12,13, rice14, rapeseed15 and tomato16. A SFT gene was reported to cause strong yield heterosis governing by over-dominance in plant architecture17. The Dw3 gene contributed to heterosis for plant height in a way of repulsion linkage in sorghum18. Additionally, novel experimental design and molecular quantitative genetics approach has been used to elucidate the importance of epistasis at two-locus level in rice during the past decades19,20,21,22. Recently, Jiang et al. suggested that dominance effects played a less prominent role than epistatic effects in grain-yield heterosis in wheat by developing a quantitative genetic framework23.
Recombinant inbred line (RIL) population is available to dissect additive and additive × additive effects but lacks heterozygous genotypes to dissect dominance and dominance-related genetic effects. So attempts have been reported by constructing testcross (TC) or backcross (BC) populations and immortalized F2 (IF2) population to create heterozygotes in rice10,14,24,25, maize11,13,26 and cotton27,28. Dominance complementation was considered as the major genetic basis of heterosis in rice because heterozygotes were superior to respective homozygotes in a BC1F1 population in rice10. Most QTLs underlying grain yield displayed apparent over-dominance effects, and little difference was observed between heterozygous genotypes of nine families of hybrids in three RIL populations in maize13. Epistasis and over-dominance were the major genetic bases of inbreeding depression and heterosis for grain and biomass yield by using five rice populations14. Heterotic effects and dominance × dominance interaction explained the genetic basis of heterosis in an IF2 population deriving from an elite rice hybrid21. Over-dominance, pseudo-over-dominance and epistasis were estimated as important contributors to yield heterosis using a high-density genetic map in rice22. Among main-effect QTLs and digenic epistatic QTLs pairs, over-dominant loci were the most important than additive, complete and partially dominant loci in two BC populations based on one same RIL population in rice24. Dominance, over-dominance and epistasis contributed to the genetic basis of heterosis using a 3,184 bin-map in an IF2 population in maize29. Moreover, new strategy of heterotic haplotype capture was proposed to trace novel heterozygous chromosome blocks for breeding30. A recent report proved that the new statistical models of QTL mapping can completely dissect large-scale time course data in post-genome era31.
Yield potential has always been a vital target of plant breeding in cotton. Significant yield heterosis was previously reported in cotton27. It is also a major breeding solution to exploit heterosis for improving yield on Upland cotton. For decades, 271 QTLs were available for yield and yield-component traits in the CottonGen database32. Among 4268 QTLs in Cotton QTLdb database33, 87, 59, 98, 169 and 305 QTLs were detected for seed-cotton yield, lint yield, boll number per plant, boll weight and lint percentage, respectively. However, less QTL have been resolved for seed-cotton yield, lint yield and boll number per plant due to complex experiment management, heavy workload and highly accurate data. The qSCYchr07a displayed strong over-dominance effect and the qSCYchr07c explained 38.96% of phenotypic variation for seed-cotton yield34. A total of 14 QTLs were identified for seed-cotton yield, lint-cotton yield and lint percentage in a RIL population of Upland cotton35. Dominance and over-dominance contributed to seed-cotton yield heterosis in an IF2 population derived from a heterotic hybrid of ‘XZM 2’ in Upland cotton36. Heterotic QTL analysis suggested that over-dominance mainly contributed to cotton yield heterosis37. Twenty-three QTLs were identified for boll weight and lint percentage in an intraspecific population of Upland cotton38. Fifty-eight QTLs were resolved for three yield-component traits but not for direct yield traits by a linkage map harboring 2618 polymorphic SNP markers39. Using two parental BC populations, 58 QTLs were also just mapped for three yield-components in Upland cotton28. Therefore, more QTLs controlling yield traits directly need to be identified and the genetic basis for yield heterosis need to be explored in Upland cotton.
In our lab, we have resolved QTL analysis and heterosis for yield and yield-components using F2, RIL and maternal backcross (BC/M) populations derived from a commercial hybrid ‘Xinza 1’ in Upland cotton. Partial dominance, over-dominance, epistasis and QTL × environment interaction contributed to yield heterosis in the three populations derived from ‘Xinza 1’27,40,41. However, no paternal backcross (BC/P) population had been used to explore the genetic basis of yield heterosis in Upland cotton. Here, we generated a total of 354 BCF1 crosses for BC/P and BC/M populations by backcrossing the 177 RI lines to GX1135 and GX100-2, respectively. Backcrossing field trials were carried out including 354 BCF1 crosses, the RI lines as current female parents and the common male parent. This experimental design has the obvious advantages: (I) dissecting all genetic components concerning dominance, over-dominance and epistasis effects, and effects by paternal and maternal parents; (II) verifying common even stable QTLs for important traits using three corresponding populations (BC/P, BC/M and RIL) originated from the same hybrid; and (III) generating enough hybrid seeds when needed, similar to IF2 population20. Seven field trials were performed across two years following a randomized complete block design with two replications. We collected phenotypic data in three corresponding populations for yield and yield-component traits. The study provides new resource to explore the genetic basis of yield heterosis in Upland cotton.
Results
Phenotypic performance of parents and populations
Table 1 presents the measurement of yield and yield components over 2015 and 2016. The original female parent GX1135 showed superior performance than the original male parent GX100-2 across multiple environments. We estimated heterosis of the hybrids on average across all environments. Seed-cotton yield (SY) and lint yield (LY) displayed 24.47% and 27.18% mid-parent hybrid vigor on average, respectively, following 10.83% for boll number per plant (BNP), 3.98% for boll weight (BW) and 3.08% for lint percentage (LP) in seven experiments. Mean values were always larger for SY, LY, BNP and LP in BC/M population than in BC/P and RIL populations in both 2015E2 and 2016E2. However, mid-parent heterosis values decreased for a same trait in BC/M population in comparison with that in BC/P population. BNP showed significant and high correlation with SY, as same as with LY (Table 2). We also estimated correlations between measurements of the same trait between the BC/M and BC/P populations in Table 2. The same trait correlated lightly or no significantly between BC/M and BC/P populations. The high correlation showed between RIL-M and RIL-P populations, validating the accuracy of the measurement. The ANOVA analysis indicated that majority of genotype variance were significant at 0.01 or 0.05 probability levels for five traits in BC/P, RIL-P, BC/M and RIL-M populations (Table 3). On the contrary, genotype × environment variance displayed non-significant difference. Heritability of SY decreased from 0.76 in RIL population and 0.64 in BC population to 0.42 in MPH-P dataset in BC/P field trials, similar tendency for majority of traits in BC/P and BC/M trials (Table 3). In addition, significantly positive correlations were observed for yield and yield-components traits between BC and MPH datasets as well as between RIL and BC datasets (Table S1). On the contrary, there was no correlation for five traits between RIL and MPH datasets.
Single-locus QTLs for yield and yield-component traits
Figure S1 and Table S2 present single-locus QTLs in multiple populations over 2 years by the composite interval mapping (CIM) method. We identified 35 QTLs in BC/M and BC/P populations, 27 heterotic loci by MPH-M and MPH-P datasets, and 41 QTLs in RIL-M and RIL-P populations.
For seed-cotton yield per plant, a total of 10 QTLs were anchored to six chromosomes, respectively. Six common and stable QTLs were identified across multiple environments or in multiple populations. The common qSY-Chr2-1 was simultaneously identified in RIL-P, BC/P and MPH-P datasets over two years. qSY-Chr2-1 explained 27.26% of phenotypic variation in BC/P population in 2016E1 and it was 12.41% in MPH-P dataset. The qSY-Chr20-1 was detected in RIL population in two continuous years. Both qSY-Chr21-1 and qSY-Chr21-2 shared between TC and RIL population.
For lint yield per plant, 13 QTLs were identified. They located on nine chromosomes. Six common QTLs explained 5.44–19.61% of phenotypic variation. Four, two and six QTLs were detected in the RIL-M, BC/M and MPH-M datasets, respectively. Six, four, four QTLs were resolved in the RIL-P, BC/P and MPH-P datasets, respectively. qLY-Chr2-1 was detected not only in RIL population across three environments but also in the BC/P and MPH-P across three environments. The QTL explained 7.75 to 19.42% of phenotypic variation. In BC/P population, both qLY-Chr2-1 and qLY-Chr2-2 displayed over-dominant effect in 2016E1 or 2016E2. At the same time, qLY-Chr2-2 contributed to lint yield heterosis, providing alleles by the female parent among RI lines. Over-dominant qLY-Chr13-1 was identified in both BC/M and MPH-M datasets. The qLY-Chr13-1 increased the lint yield with negative additive effect.
For boll number per plant, 23 QTLs were detected on 13 different chromosomes. There are seven common QTLs across multiple environments or datasets. Three QTLs (qBNP-Chr1-3, qBNP-Chr2-2, and qBNP-Chr14-2) were repeatedly identified in RIL population across more than one environment. Two common QTLs (qBNP-Chr15-1, qBNP-Chr21-1) validated each other in RIL and either of two BC populations. And qBNP-Chr21-2 explained 8.88% and 12.22% of phenotypic variation in BC/M population in 2015E1 and 2016E2, respectively. Two common heterotic loci (qBNP-Chr1-4 and qBNP-Chr15-1) were detected only in BC/P population. qBNP-Chr1-4 displayed apparent over-dominant effect with d/a = 2.60 and qBNP-Chr15-1 increased one boll number.
Here, nine QTLs were identified for boll weight. Six common QTLs located on chromosome 2, 5, 6, 20 and 23 across more than one environment, respectively. The common QTLs also showed same genetic effect orientation. The stable qBW-Chr5-2 was identified in RIL population for four times across three locations in two years. It improved boll weight with partial dominance effect in both BC populations. Four heterotic QTLs (qBW-Chr5-1, qBW-Chr5-2, qBW-Chr6-1 and qBW-Chr23-1) shared in BC/M and BC/P populations. The four QTLs explained 9.54%, 10.73%, 6.19% and 5.54% of phenotypic variation on average, respectively. The common QTL qBW-Chr20-1 increased boll weight in both RIL and BC/P populations. The QTL provided alleles with negative additive effects.
For lint percentage, seven, six and three QTLs were resolved in RIL-M, BC/M and MPH-M datasets, respectively. Then, seven, five and four QTLs were identified in RIL-P, BC/P and MPH-P datasets, respectively. Three QTLs (qLP-Chr5-1, qLP-Chr5-2 and qLP-Chr13-3) verified in RIL, BC/M and BC/P populations at the same time. The qLP-Chr5-2 was simultaneously detected in the BC/M, BC/P and RIL populations. The qLP-Chr19-1 and qLP-Chr13-2 were found only in BC/P population, while qLP-Chr4-1, qLP-Chr7-1 and qLP-Chr13-2 were observed just in BC/M population. All of the five common QTLs were also detected in RIL population.
Taken together, 71 QTLs were detected for five yield and yield-component traits, including 35 common QTLs (49.30%) in more than one environment or population. A total of 21 QTLs were detected only in BC/M population and 20 QTLs only in BC/P population. Six QTLs were simultaneously detected in both BC/M and BC/P populations. In addition, 12 and 15 heterotic loci were identified using MPH-M and MPH-P datasets, respectively. But there is no common heterotic locus in both MPH datasets. However, 11 common heterotic loci overlapped with seven QTLs in BC/P or RIL-P population and four QTLs in BC/M or RIL-M population (Fig. S1). These overlapping regions distributed on chromosome 1, 2, 5, 6, 7, 13, 15, 21 and 23, respectively.
Genetic effect at single locus level
Three types of genetic effects were summarized for single locus QTLs in two BC populations and two MPH datasets (Table 4). In BC/M population, 19 (51.35%) additive QTLs and 12 (32.43%) over-dominant QTLs contributed much to heterosis, following six (16.22%) partial dominant QTLs. Homozygous P2P2 recessive alleles providing by paternal parent changed to be heterozygous P1P2 alleles in RIL population after crossing to GX1135. In BC/P population, ten (31.25%) additive QTLs and six (18.75%) partial dominant QTLs played slight role in performance than 16 (50.00%) over-dominant QTLs. Homozygous P1P1 dominant alleles from maternal parent changed to be heterozygous P1P2 alleles in RIL population after crossing to GX100-2. In BC/M population, there was more over-dominant QTLs for LY, whereas more additive QTLs was resolved for BNP and BW. However, we detected the most over-dominant QTLs for SY and LY in BC/P population. In addition, additive, partial dominant and over-dominant QTLs played important role together for LP in both BC populations.
Relationship between whole-genome marker heterozygosity and performance
The experimental design allowed us to dissect relationships between whole-genome marker heterozygosity and trait performance in BC/M, BC/P, MPH-M, and MPH-P datasets. We examined correlations between whole-genome marker heterozygosity of 654 polymorphic loci and one phenotypic dataset for yield and yield-component traits. The majority of correlations showed no significance in all of BC/M, BC/P, MPH-M, or MPH-P datasets (Table S3), demonstrating that overall marker heterozygosity contributed little to yield heterosis. The result was consistent with previous reports10,20,27. A previous study demonstrated that a few loci from female parents explained a large proportion of the yield advantage of hybrids but not universally via integrated genomic analyses42.
Pleiotropic region and genetic contribution
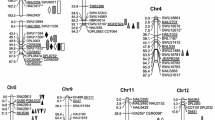
Totally, 16 clusters showed pleiotropic effects in 20 cM interval. These clusters involved in 45 QTLs (63.38%) in present study (Fig. S1, Table S4). Cluster-Chr2-1 of SWU11889-SWU11976 was detected in BC/P and MPH-P datasets. The cluster increased 4.30 g seed-cotton yield, 2.26 g lint yield, 1 boll per plant and 0.11 g boll weight. Cluster-Chr5-2 and Cluster-Chr21-1 controlled seed-cotton yield, lint yield and yield-components at the same time.
Six QTL clusters contained QTLs for seed-cotton yield and lint yield at the same time. Importantly, four stable pleiotropic heterotic regions simultaneously controlled seed-cotton yield and lint yield on chromosome (Chr) 2, Chr 5, Chr 11 and Chr 25. Interestingly, all of heterotic loci showed over-dominant effect in the four stable pleiotropic heterotic regions. In BC/P populations, qSY-Chr2-1 and qLY-Chr2-1 clustered together and increased yield providing alleles with positive effect value. Three pleiotropic regions increased yield with negative effect value, harboring over-dominant qSY-Chr5-2 and qLY-Chr5-1, qSY-Chr11-1 and qLY-Chr11-1, qSY-Chr25-1 and qLY-Chr25-1, respectively. We searched for physical locations of the flanking markers based on the CottonFGD database43. The qBNP-Chr5-2 in BC/M population and the common QTL qLP-Chr5-3 in RIL population shared a 333 kb pleiotropic region (Table S4). The pleiotropic region of TMB1296-HAU1603 contained 48 genes on chromosome 5 in Upland cotton (Table S5). Another 516 kb pleiotropic region of NAU2152-NAU5428 contained 25 genes including qSY-Chr11-1 and qLY-Chr11-1. The qSY-Chr11-1 and qLY-Chr11-1 showed over-dominant effect.
Epistasis across multiple environments
We estimated QTLs by inclusive composite interval mapping (ICIM) method in RIL-M, BC/M and MPH-M datasets, respectively. A total of 92, 60 and 35 main effect QTL and QTL × environments interaction (M-QTLs and QEs) were detected for five yield and yield-component traits (Tables 5, S6-A, S6-C, S6-E). The M-QTLs explained 0.41 to 2.47% of phenotypic variation (PV), while the QEs explained 0.14 to 1.49% of PV. At two-locus level, 314, 75 and 36 epistasis QTLs (E-QTLs and QQEs) were identified in RIL-M, BC/M and MPH-M datasets, respectively. The E-QTLs explained 1.06 to 2.93% of PV and QQEs explained 0.58 to 1.48% of PV (Tables 5, S6-B, S6-D, and S6-F).
A total of 57, 34 and 22 M-QTLs and QEs were detected for five yield and yield-component traits in RIL-P, BC/P and MPH-P datasets across three environments, respectively (Tables 5, S7-A, S7-C, S7-E). On average, the M-QTLs explained 1.86% to 2.89% of PV while the QEs explained 0.24% to 0.75% of PV. At two-locus level, we detected 115, 57 and 21 E-QTLs and QQEs in RIL-P, BC/P and MPH-P datasets, respectively. The E-QTLs explained 0.65 to 3.96% of PV and QQEs explained 0.14% to 2.80% of PV (Tables 5, S7-B, S7-D, and S7-F).
Three genetic types of epistasis were resolved for yield and yield-component traits in RIL-M, RIL-P, BC/M, BC/P, MPH-M and MPH-P datasets (Table S8): Type I, interaction between two M-QTLs; Type II, interaction between one M-QTL and non M-QTL; Type III, interaction between one non M-QTL and another non M-QTL27. The number of E-QTLs with Type III was 241, 59, 27 in RIL-M, BC/M and MPH-M datasets, respectively; that with Type II is 70, 14, 8; that with Type I is 3, 2, 1. The numbers of E-QTLs were 2, 0, 0 for Type I, respectively; 17, 4, 4 for Type II; 93, 57, 13 for Type III. The result demonstrated that large number of E-QTLs contributed to phenotype in RIL and BC populations and heterosis between numerous non M-QTLs, which displayed minor effect together.
Discussions
Experimental design for heterotic loci by multiple mapping populations
The extended design of NCIII was suitable to explore heterosis by creating heterozygotes by backcrossing or testcrossing parental lines to RIL or doubled haploid (DH) lines25,27,28. In the present study, we constructed two parental BC populations based on one RIL population as the permanent experimental design. Superior phenotypic values displayed in BC/M population rather than that in BC/P population, suggesting that the performance of both parents determined the performance of their hybrid. In all of seven field trials, every backcrossing progeny inter-planted in the middle of both parents. The experimental design allowed calculating the mid-parent heterosis (MPH) so as to detect heterotic loci for measuring heterotic effect directly24. Hua et al. separated 33 heterotic loci that caused yield heterosis in rice21. Here, 27 heterotic loci were resolved from two parental BC populations (Table S2). Two stable heterotic loci (qSY-Chr2-1 and qLY-Chr2-1) shared the region of SWU11889-SWU11950. The qSY-Chr2-1 increased 2.61 g – 8.72 g seed-cotton yield across four environments (Table S9). At the same time, qSY-Chr2-1 explained 12.85% and 27.26% of phenotypic variation in MPH-P and BC/P datasets, respectively. The qLY-Chr2-1 increased 1.36 g- 4.11 g lint yield across six environments in 2015, 2016 and previous 201227,41 (Table S9). In addition, the QTL explained 12.41% and 19.61% of phenotypic variation in the MPH-P and BC/P datasets, respectively. Both qSY-Chr2-1 and qLY-Chr2-1 displayed apparent over-dominance effect (OD) with the degree of dominance of d/a ranged from 1.6 to 4.9 in BC/P population. Three other common and clustering heterotic loci contributed to yield heterosis by the same genetic mode (Fig. S1, Table 5). These heterotic loci shared the region controlling SY and LY on chromosome 5, 11 and 25, respectively. Interestingly, 11 heterotic loci (64.71%) mentioned above overlapped with the QTLs which were detected in both BC/M and BC/P populations for yield-component traits, including qSY-Chr2-1, qLY-Chr2-1, qLY-Chr2-2, qLY-Chr2-3, qLY-Chr13-1, qBNP-Chr1-4, qBW-Chr5-1, qBW-Chr23-1, qLP-Chr5-2, qLP-Chr7-1 and qLP-Chr13-3. The results implied that some heterotic loci linked with QTLs together among five yield and yield-component traits. However, only two common heterotic loci (2.82%) were identified across multiple environments or populations in present study, including qSY-Chr2-1 and qLY-Chr2-1. The result assumed that each measurement depended on the neighboring materials in one plot because of the sensitivity to environment for yield heterosis and the apparent marginal effect of Upland cotton plant.
Common QTLs and their genetic effects at single locus level
In present study, 35 common QTLs (49.30%) were identified in more than one environment or population by using RIL, BC/M, BC/P, MPH-M and MPH-P datasets. In previous study, 58 common QTLs were identified using RIL and BC/M populations27,41. Among these common QTLs, 17 major QTLs explained over 10.00% of phenotypic variation. A total of 19 QTLs in present study were same to previous QTLs in three BC/M trials in 201227 (Table S9). Totally, 9 common previous QTLs were identified in the F2 populations in 2008 and 200940 (Table S10). Three QTLs of Cluster-Chr5-1 increased boll weight and lint percentage at the same time over F2, RIL, BC/M and BC/P populations, including qBW-Chr5-1, qBW-Chr5-2 and qLP-Chr5-1 (Tables S4, S9, S10). Taken together, a total of nine common QTLs validated across multiple years of 2012, 2015 and 2016 for seed-cotton yield and lint yield traits. The region of SWU20917-NAU6240 explained 10.78–37.72% of phenotypic variation across multiple environments in RIL and BC populations. All of QTLs flanking with SWU11887 increased phenotypic performance of SY, LY and BW on chromosome 2 (Table S4). In this study, the BNL1317 flanking qSY-Chr9-1 for seed-cotton yield was common to the previous QTL with LOD 4.94 controlling lint percentage35. In present study, three markers of Gh157, BNL1495 and CGR5390 involved in qBNP-Chr13-1, qLP-Chr13-2 and qLP-Chr13-3 for boll number per plant and lint percentage on chromosome 13. The three markers were common to the previous markers for lint percentage35, the previous QTL (qLY-Chr13-1) for lint yield40 and previous association locus qLP-D5-1 for lint percentage44. There are 150 same SSR markers between our linkage map in present study and the previous map including 2051 SSR loci38. Two QTLs (qLY-Chr21-3, qBNP-Chr21-3) involving in BNL3442a increased lint yield and boll number per plant in our study, similar in another previous report36. The common QTLs and validated QTLs across multiple environments and multiple populations provide a valuable resource for MAS and the further research. The results indicated that the design in present study was efficient to map common even stable QTLs or heterotic loci across multiple populations. In addition, we observed 48 genes in a 333 kb region (TMB1296-HAU1603) in the reference genome. And the 516 kb pleiotropic region (NAU2152-NAU5428) contained 25 genes in the reference genome (Table S5). In further study, we will focus on the two regions for fine mapping and gene function analysis. The availability of cotton genomic data for diploid species45,46,47, tetraploid genomes48,49,50,51 facilitated the development of single nucleotide polymorphism (SNP) markers. Until now, 302,735 SNPs were deposited in CottonGen database32. The development of CottonSNP63K array and high-throughput genotyping arrays facilitate applications of SNP markers to linkage mapping and GWAS in cotton39,42,43,44,45,46,47,48.
Genetic basis on heterosis in Upland cotton
At single locus level, 19 (51.35%), 6 (16.22%), 12 (32.43%) QTLs were estimated in BC/M population for additive effect, partial dominance effect and over-dominance effect, respectively (Table 4). In BC/P population, the number of QTLs were 10 (31.25%) for additive effect, 6 (18.75%) for partial dominance effect and 16 (50.00%) for over-dominance effect, respectively. The result indicated that three types of genetic effects were detected at the single-locus level in BC/M population, similarly in BC/P population. However, the most QTLs showed additive effect, following over-dominant effect in BC/M population. The result was consistent with a null hypothesis that gene expression will be additive in the hybrid in comparison with their expression in the parents52. However, the most QTLs showed over-dominant effect in BC/P population. For yield and yield-component traits, additive effect is the most important in hybrids by crossing RI lines to superior performance parent harboring dominant alleles, whereas partial dominant and over-dominant effects are major genetic basis in hybrids by crossing RI lines to inferior performance parent harboring recessive alleles.
Epistasis refers to the interaction between alleles from different loci7. In present study, 75, 36, 57 and 21 epistatic QTLs (E-QTLs and QQEs) were identified in BC/M, MPH-M, BC/P and MPH-P datasets, respectively (Tables 5 and S6, S7). The result indicated that epistasis contributes to heterosis in consistent with previous studies19,20,21,22. Moreover, E-QTLs and QQE explained higher phenotypic variation (PV) than that by main effect QTL by environments (M-QTL and QEs) in both BC/M and BC/P populations. The QTLs for same trait explained more portion of PV in BC/P and MPH-P datasets than that in the BC/M and MPH-M datasets. On the contrary, QTL × environments interaction (QE or QQE) explained less PV in BC/P and MPH-P datasets than that in BC-M and MPH-M datasets. The results indicated that E-QTLs played role in yield heterosis in ‘Xinza 1’. Environment only explained 0.13–2.80% of PV at two-locus level, suggesting that yield heterosis was sensitive to environment in Upland cotton (Table 5). In a short, we detected additive, partial dominance, over-dominance at single locus level together with epistasis and environment interactions. The results indicated that cumulative effects controlled yield heterosis in Upland cotton, consistently with the previous results27,40. Guo et al. reported the contribution of over-dominant QTLs for heterosis by using an interspecific cotton population37. Similarly, genetic basis of grain yield heterosis were the cumulative effects of dominance, over-dominance, and epistasis in maize hybrid ‘Yuyu 22’29. Recently, additive, partial dominance and over-dominance controlled heterosis owing to allelic dosage effects in maize53,54 and rice42. The genome-wide heterozygosity of hybrids made a limited contribution in present study. The result was in consistent with previous report for biomass heterosis by characterizing the genomic architecture in 200 Arabidopsis hybrids55. As other polyploidy plants, cotton also exhibits better vigor after polyploidization event52. The yield and its component traits are complex quantitative traits. The genetic basis of heterosis is mysterious especially the allotetraploid Upland cotton. Pleitropic regions involving in heterotic loci contained numbers of genes. The regions with cumulative genetic effects maybe regulate yield heterosis in a particular inheritance mode such as dosage effects. Further work need to explore heterosis mechanism using one single and novel gene in Upland cotton.
Methods
Development of the experimental populations
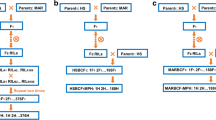
Recombinant inbred line (RIL) population were previous developed by single seed descent method, which derived from the Upland cotton hybrid ‘Xinza 1’ (GX1135 × GX100-2)27,40. The 177 F14 individuals of RIL population were re-planted for inbred seeds. A total of 354 progenies were generated by backcrossing 177 RI lines to GX1135 (as the present common male parent) and GX100-2 (as the present common male parent), respectively. We named the maternal and paternal backcross populations as BC/M and BC/P populations for short, respectively, the same as RIL-M population and MPH-M dataset in BC/M field trials, and RIL-P population and MPH-P dataset in BC/P field trials (See below).
Field design and management
Two kinds of backcross trials were carried out as follows: (I) the paternal BC trial (BC/P field trial), containing BC/P population by cross of 177 F14 RILs × GX100-2, RIL-P population and common male parent GX100-2 (original male parent); (II) the maternal BC field trial (BC/M trial), containing BC/M population by cross of 177 F14 RILs × GX1135, RIL-M population and common male parent GX1135 (original female parent). We carried out seven field trials over two years of 2015 and 2016 at three locations in China as follows: E1, Handan, Hebei Province; E2, Cangzhou, Hebei Province; E3, Wuhan, Hubei Province56. Four BC/M trials were performed in 2015E1, 2015E2, 2015E3 and 2016E2 (the year and location). Three BC/P field trials were constructed in 2015E2, 2016E1, and 2016E2. All of seven field trials followed a randomized complete block design with two replications. The control set was planted in seven field trials, respectively, including GX1135, ‘Xinza 1’ F1, GX100-2 and a competition control of Upland cotton hybrid variety (‘Ruiza 816’ or ‘Ezamian 1’)56. Unfortunately, three field trials encountered hailstone disaster on June 11, 2015 in E2 and on June 28, 2016 in E1. After the hailstone disasters, we immediately performed effective field managements to recover the damaged plants (2015E2 for one BC/P trial and one BC/M trial, 2016E3 for one BC/P trial). Therefore, we regarded as the experiments in a same identical and natural environment because of the well recovery of the plants (Fig. S2). The details for three field trials at E1 and E2 in 2016 are same to the arrangement in 2015 in the previous report56. The field management followed the conventional standard field practices.
Phenotypic evaluation
We scored eight plants except the marginal one for phenotypic performance. We harvested seed cotton in each plot for seed-cotton yield per plant (SY) and boll number per plant (BNP) at maturity stage. Twenty-five naturally opening bolls were randomly hand-harvested in the middle of plants for boll weight (BW) and lint percentage (LP). We evaluated lint yield per plant (LY) by multiplying SY by LP. In addition, SY in 2015E2 was predicted by multiplying BNP and BW due to unfavorable hailstone disasters. A total of 45,174 plants were measured for yield and yield-component traits at three locations across two testing years. At last, we collected seven complete datasets from four BC/M trials and three BC/P trials for five yield and yield-component traits.
The genetic linkage map information
A total of 623 polymorphic SSR loci were previously classified into 31 linkage groups anchored on 26 chromosomes27. The genetic map covered 3889.9 cM (88.20%) with interval of 6.2 cM on average. The genotypes of 177 individuals in the RIL population were reported previously, as well as that of the BC/M population27. The genotypic data of the BC/P population were deduced from that of RIL population (Table S11).
Statistical analysis
Data for five yield and yield-component traits were obtained over 2 years from four populations (the RIL-M, BC/M, RIL-P, BC/P population) and the currently common male parents. Basic statistical analysis was performed by the software SPSS (version 19.0, SPSS, Chicago). Two mid-parents heterosis datasets (MPH-M and MPH-P datasets) were assessed by the equation:
where F1 refers to phenotype value of each BC1F1 progeny in BC/M or BC/P populations; P1 refers to the recurrent female parent of the RIL-M or RIL-P populations; P2 refers to the currently male donor parent of GX1135 in BC/M trial or GX100-2 in BC/P trial. Heterosis (%) was assessed by the equation30
The dataset of mean value for two replications were used to map quantitative trait locus (QTL) in every single environment for five yield-related traits. We estimated variance for multiple datasets in multiple environments for five yield-related traits by R software. The linear model formula was as
where G refers to genotype effect, E to environment effect, G × E to genotype-by-environment interaction effect, block to repeat effects in one environment, error to error effect. Based on the variance component, heritability was calculated in the equation as
where δ2G, δ2G×E, and δ2e refer to the genotypic variance, genotype-by-environment interaction variance, and error variance, env to the number of the environments, and rep to the number of replications per environment57. For the low-accuracy raw datasets, larger error variance was not allowed to estimate heritability due to the environment sensitivity and/or bigger artificial error. We used QTL Cartographer (Version 2.5)58 to map single-locus QTL by composite interval mapping (CIM) method. The genetic effect values were estimated in the confidence interval of 95%. The threshold of LOD values were estimated after 1000 permutations tests to declare a significant QTL with a significant level of P < 0.05. However, a common QTL was considered with LOD 2.0 in another environment or population27,39. Common QTLs were evaluated by linked position and shared common markers59. Stable QTLs in the present study referred to common QTLs with stable genetic effect orientation in multiple environments and/or populations. The degree of dominance was estimated for common QTLs derived from different populations or datasets15. Genetic effects of single-locus QTLs were defined as: additive effect loci just detected in BC populations, the complete or partial dominance effect loci with d/a ≤ 1, over-dominance effect loci with d/a > 1 or QTLs detected by MPH dataset25,60. The genetic effects of single QTL were assessed following: additive effect,
where P1P1 and P2P2 stands for effects of homozygous genotypes from RIL population; dominance effect,
where P1P2 and P2P2 stand for effects of heterozygous genotypes after correction of BC1F14 observation for mid-parent performance (MPH);
for BC/M and BC/P populations25,27. The software IciMapping 4.1 (www.isbreeding.net) was used to test additive, dominance and epistasis under environments by inclusive composite interval mapping (ICIM) method. The main-effect QTL (M-QTL) and its environmental interaction (QTL × environment, QE), epistatic QTLs (E-QTLs) and its environmental interactions (QTLs × environment, QQE) were conducted using RIL, BC and MPH datasets under multiple environments in two parental BC trials. A threshold LOD 2.5 and 5 scores were used to declare significant M-QTL and E-QTLs, respectively27,61,62,63,64,65,66.
Data Availability
The authors state that all data necessary for confirming the conclusions presented in the article represented fully within the article and in the Table S11.
References
Bruce, A. The Mendelian theory of heredity and the augmentation of vigor. Science 32, 627–628 (1910).
Jones, D. F. Dominance of linked factors as a means of accounting for heterosis. Genetics 2, 466–479 (1917).
Shull, G. H. The composition of a field of maize. J. Hered. 4, 296–301 (1908).
East, E. Heterosis. Genetics 21, 375–397 (1936).
Crow, J. F. Dominance and over-dominance. in Heterosis 282–297 (Iowa State College Press, Ames, Iowa, USA (1952).
Schnable, P. S. & Springer, N. M. Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 64, 71–88 (2013).
Richey, F. D. Mock-dominance and hybrid vigor. Science 96, 280–281 (1942).
Minvielle, F. Dominance is not necessary for heterosis: a two-locus model. Genet. Res. 49, 245–247 (1987).
Schnell, F. W. & Cockerham, C. C. Multiplicative vs. arbitrary gene action in heterosis. Genetics 131, 461–469 (1992).
Xiao, J. H., Li, J. M., Yuan, L. P. & Tanksley, S. D. Dominance is the major genetic-basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140, 745–754 (1995).
Frascaroli, E. et al. Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics 176, 625–644 (2007).
Stuber, C. W. et al. Identification of genetic-factors contributing to heterosis in a hybrid from 2 elite maize inbred lines using molecular markers. Genetics 132, 823–839 (1992).
Lariépe, A. et al. The genetic basis of heterosis: multiparental quantitative trait loci mapping reveals contrasted levels of apparent over-dominance among traits of agronomical interest in maize (Zea mays L.). Genetics 190, 795–811 (2012).
Li, Z. K. et al. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158, 1737–1753 (2001).
Radoev, M., Becker, H. C. & Ecke, W. Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179, 1547–1558 (2008).
Semel, Y. et al. Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. USA 103, 12981–12986 (2006).
Krieger, U., Lippman, Z. B. & Zamir, D. The flowering gene single flower truss drives heterosis for yield in tomato. Nat. Genet. 42, 138–459 (2010).
Li, X., Fridman, E., Tesso, T. T. & Yu, J. Dissecting repulsion linkage in the dwarfing gene Dw3 region for sorghum plant height provides insights into heterosis. Proc. Natl. Acad. Sci. USA 112, 11823–11828 (2015).
Yu, S. et al. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 94, 9226–9231 (1997).
Hua, J. et al. Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 162, 1885–1895 (2002).
Hua, J. et al. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 100, 2574–2579 (2003).
Zhou, G. et al. Genetic composition of yield heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 109, 15847–15852 (2012).
Jiang, Y., Schmidt, R. H., Zhao, Y. & Reif, J. C. A quantitative genetic framework highlights the role of epistatic effects for grain-yield heterosis in bread wheat. Nat. Genet. 49, 1741–1746 (2017).
Mei, H. W. et al. Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two backcross populations. Theor. Appl. Genet. 110, 649–659 (2005).
Luo, X. et al. Additive and over-dominant effects resulting from epistatic loci are the primary genetic basis of heterosis in rice. J. Integr. Plant Biol. 51, 393–408 (2009).
Tang, J. et al. Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor. Appl. Genet. 120, 333–340 (2010).
Shang, L. et al. Epistasis together with partial dominance, over-dominance and QTL by environment interactions contribute to yield heterosis in upland cotton. Theor. Appl. Genet. 129, 1429–1446 (2016).
Wang, H. T. et al. Identification of QTL for fiber quality and yield traits using two immortalized backcross populations in upland cotton. PloS ONE 11, e0166970 (2016).
Guo, T. et al. Genetic basis of grain yield heterosis in an “immortalized F2” maize population. Theor. Appl. Genet. 127, 2149–2158 (2014).
Snowdon, R. J. et al. Heterotic Haplotype Capture: precision breeding for hybrid performance. Trends Plant Sci. 20, 410–413 (2015).
Li, Z. & Sillanpää, M. J. Dynamic quantitative trait locus analysis of plant phenomic data. Trends Plant Sci. 20, 822–833 (2015).
Yu, J. et al. Cottongen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, 1229–1236 (2014).
Said, J. I., Knapka, J. A., Song, M. Z. & Zhang, J. F. Cotton QTLdb: a cotton QTL database for QTL analysis, visualization, and comparison between Gossypium hirsutum and G. hirsutum × G. barbadense populations. Mol. Genet. Genomics 290, 1615–1625 (2015).
He, D. et al. QTL mapping for economic traits based on a dense genetic map of cotton with PCR-based markers using the interspecific cross of Gossypium hirsutum × Gossypium barbadense. Euphytica 153, 181–197 (2007).
Wu, J. et al. Quantitative analysis and QTL mapping for agronomic and fiber traits in an RI population of upland cotton. Euphytica 165, 231–245 (2009).
Liu, R. et al. Quantitative trait loci mapping for yield and its components by using two immortalized populations of a heterotic hybrid in Gossypium hirsutum L. Mol. Breeding 29, 297–311 (2012).
Guo, X. et al. Mapping heterotic loci for yield and agronomic traits using chromosome segment introgression lines in cotton. J. Integr. Plant Biol. 55, 759–774 (2013).
Liu, X. et al. Enriching an intraspecific genetic map and identifying QTL for fiber quality and yield component traits across multiple environments in Upland cotton (Gossypium hirsutum L.). Mol. Genet. Genomics 292, 1281–1306 (2017).
Li, C. et al. Genome-wide SNP linkage mapping and QTL analysis for fiber quality and yield traits in the Upland cotton recombinant inbred lines population. Front. Plant Sci. 7, 1356 (2016).
Liang, Q., Shang, L., Wang, Y. & Hua, J. Partial dominance, over-dominance and epistasis as the genetic basis of heterosis in upland cotton. PLoS ONE 10, e0143548 (2015).
Shang, L. et al. Partial dominance, over-dominance, epistasis and QTL by environment interactions contribute to the heterosis in two upland cotton hybrids. G3-Genes Genom. Genet. 6, 499–507 (2016).
Huang, X. et al. Genomic architecture of heterosis for yield traits in rice. Nature 537, 629–633 (2016).
Zhu, T. et al. CottonFGD: an integrated functional genomics database for cotton. BMC Plant Biol. 17, 101 (2017).
Zhang, T. et al. Variations and transmission of QTL alleles for yield and fiber qualities in upland cotton cultivars developed in China. PloS ONE 8, e57220 (2013).
Paterson, A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427 (2012).
Wang, K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103 (2012).
Li, F. et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572 (2014).
Zhang, T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537 (2015).
Li, F. et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530 (2015).
Liu, X. et al. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. UK 5, 14139 (2015).
Yuan, D. et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. UK 5, 17662 (2015).
Birchler, J. A., Yao, H., Chudalayandi, S., Vaiman, D. & Veitia, R. A. Heterosis. Plant Cell 22, 2105–2112 (2010).
Yao, H., Dogra, G. A., Auger, D. L. & Birchler, J. A. Genomic dosage effects on heterosis in triploid maize. Proc. Natl. Acad. Sci. USA 110, 2665–2669 (2013).
Birchler, J. A., Johnson, A. F. & Veitia, R. A. Kinetics genetics: Incorporating the concept of genomic balance into an understanding of quantitative traits. Plant Sci. 245, 128–134 (2016).
Yang, M. et al. Genomic architecture of biomass heterosis in Arabidopsis. Proc. Natl. Acad. Sci. USA 114, 8101–8106 (2017).
Ma, L. et al. QTLs analysis and validation for fiber quality traits using maternal backcross population in Upland cotton. Front. Plant Sci. 8, 2168 (2017).
Liu, W. et al. Potential for marker-assisted simultaneous improvement of grain and biomass yield in triticale. Bioenergy Research 10, 449–455 (2016).
Wang, S., Basten, C. & Zeng, Z. Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC (2012).
Shao, Q. et al. Identifying QTL for fiber quality traits with three upland cotton (Gossypium hirsutum L.) populations. Euphytica 198, 43–58 (2014).
Melchinger, A. E., Utz, H. F. & Schön, C. C. Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics 149, 383–403 (1998).
Su, J. et al. Identification of favorable SNP alleles and candidate genes for traits related to early maturity via GWAS in upland cotton. BMC Genomics 17, 687 (2016).
Bazakos, C. et al. New strategies and tools in quantitative genetics: how to go from the phenotype to the genotype. Annu. Rev. Plant Biol. 68, 435–455 (2017).
Fang, L. et al. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 49(7), 1089–1098 (2017).
Qi., H. et al. Construction of a high-density genetic map using genotyping by sequencing (GBS) for quantitative trait loci (QTL) analysis of three plant morphological traits in upland cotton (Gossypium hirsutum L.). Euphytica 213, 83 (2017).
Zhang, Z. et al. Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to Quantitative Trait Loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 16, 79 (2016).
Zhang, Z. et al. Construction of a high-density genetic map and its application to QTL identification for fiber strength in upland cotton. Crop Sci. 57, 774–788 (2017).
Acknowledgements
We thank Hanqing Liu of China Agricultural University, Xiaofang Yin, Cuicui Zuo, Dongyong Xu, and Huaiyu Lu of Guoxin Seed Company Ltd, Cangzhou, Hebei Province for their contributions on field experiments and data acquisition. Thanks to Dr. Lianguang Shang, Dr. Qingzhi Liang, Ms. Xiaocui Wang, Mr. Abdugheni Abduwelli, Ms Yuhua Li (China Agricultural University) for their contributions in molecular marker analysis and map construction. Thanks to Prof. Dr. Kunbo Wang and Fang Liu (Institute of Cotton Research, Chinese Academy of Agricultural Sciences), Prof. Dr. Zhengsheng Zhang (Southwest University) for providing parts primers of SSR markers. Thanks to Dr. Tamara L. Church (Department of Materials and Environmental Chemistry, Stockholm University) for her revision on the Introduction. This research was supported by National Key R & D Program for Crop Breeding (2016YFD0101407) to J. Hua.
Author information
Authors and Affiliations
Contributions
L.M. performed the experiments, analyzed the data and prepared the manuscript. Y.W. maintained the experimental platform and attended bench work. B.I. attended part of the experiments and discussion. J.H. conceived the experiments, provided experimental platform and revised the manuscript. All the authors approved the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ma, L., Wang, Y., Ijaz, B. et al. Cumulative and different genetic effects contributed to yield heterosis using maternal and paternal backcross populations in Upland cotton. Sci Rep 9, 3984 (2019). https://doi.org/10.1038/s41598-019-40611-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-40611-9
- Springer Nature Limited
This article is cited by
-
Molecular mapping of QTLs for fiber quality traits in Gossypium hirsutum multi-parent recombinant inbred lines
Euphytica (2021)
-
QTL controlling fiber quality traits under salt stress in upland cotton (Gossypium hirsutum L.)
Theoretical and Applied Genetics (2021)
-
QTL and genetic analysis controlling fiber quality traits using paternal backcross population in upland cotton
Journal of Cotton Research (2020)