Abstract
Molecular conjugation refers to methods used in biomedicine, advanced materials and nanotechnology to link two partners — from small molecules to large and sometimes functionally complex biopolymers. The methods ideally have a broad structural scope, proceed under very mild conditions (including in H2O), occur at a rapid rate and in quantitative yield with no by-products, enable bioorthogonal reactivity and have zero toxicity. Over the past two decades, the field of click chemistry has emerged to afford us new and efficient methods of molecular conjugation. These methods are based on chemical reactions that produce permanently linked conjugates, and we refer to this field here as covalent click chemistry. Alternatively, if molecular conjugation is undertaken using a pair of complementary molecular recognition partners that associate strongly and selectively to form a thermodynamically stable non-covalent complex, then we refer to this strategy as non-covalent click chemistry. This Perspective is concerned with this latter approach and highlights two distinct applications of non-covalent click chemistry in molecular conjugation: the pre-assembly of molecular conjugates or surface-coated nanoparticles and the in situ capture of tagged biomolecular targets for imaging or analysis.



Similar content being viewed by others
References
Janaratne, T. K., Okach, L., Brock, A. & Lesley, S. A. Solubilization of native integral membrane proteins in aqueous buffer by noncovalent chelation with monomethoxy polyethylene glycol (mPEG) polymers. Bioconjug. Chem. 22, 1513–1518 (2011).
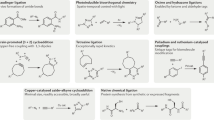
Kolb, H. C., Finn, M. G. & Sharpless, K. B. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 40, 2004–2021 (2001).
Xu, Z., Wang, L., Fang, F., Fu, Y. & Yin, Z. A review on colloidal self-assembly and their applications. Curr. Nanosci. 12, 725–746 (2016).
Plutschack, M. B., Pieber, B., Gilmore, K. & Seeberger, P. H. The hitchhiker’s guide to flow chemistry. Chem. Rev. 117, 11796–11893 (2017).
Liu, K. et al. Molecular imaging probe development using microfluidics. Curr. Org. Synth. 8, 473–487 (2011).
Cheng, Z., Al Zaki, A., Hui, J. Z., Muzykantov, V. R. & Tsourkas, A. Multifunctional nanoparticles: cost versus benefit of adding targeting and imaging capabilities. Science 338, 903–910 (2012).
Marqués-Gallego, P. & de Kroon, A. I. P. M. Ligation strategies for targeting liposomal nanocarriers. Biomed Res. Int. 2014, 129458 (2014).
Abd Ellah, N. H. & Abouelmagd, S. A. Surface functionalization of polymeric nanoparticles for tumor drug delivery: approaches and challenges. Expert Opin. Drug Deliv. 14, 201–214 (2017).
Stéen, E. J. L. et al. Pretargeting in nuclear imaging and radionuclide therapy: improving efficacy of theranostics and nanomedicines. Biomaterials 179, 209–245 (2018).
Yoon, H. Y., Koo, H., Kim, K. & Kwon, I. C. Molecular imaging based on metabolic glycoengineering and bioorthogonal click chemistry. Biomaterials 132, 28–36 (2017).
Rossin, R. & Robillard, M. S. Pretargeted imaging using bioorthogonal chemistry in mice. Curr. Opin. Chem. Biol. 21, 161–169 (2014).
Patterson, D. M., Nazarova, L. A. & Prescher, J. A. Finding the right (bioorthogonal) chemistry. ACS Chem. Biol. 9, 592–605 (2014).
Freidel, C., Kaloyanova, S. & Peneva, K. Chemical tags for site-specific fluorescent labeling of biomolecules. Amino Acids 48, 1357–1372 (2016).
Rashidian, M., Dozier, J. K. & Distefano, M. D. Enzymatic labeling of proteins: techniques and approaches. Bioconjug. Chem. 24, 1277–1294 (2013).
Dundas, C. M., Demonte, D. & Park, S. Streptavidin–biotin technology: improvements and innovations in chemical and biological applications. Appl. Microbiol. Biotechnol. 97, 9343–9353 (2013).
Jain, A. & Cheng, K. The principles and applications of avidin-based nanoparticles in drug delivery and diagnosis. J. Control. Release 245, 27–40 (2017).
Miller, L. W., Cai, Y., Sheetz, M. P. & Cornish, V. W. In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat. Methods 2, 255–257 (2005).
Xu, S. & Hu, H.-Y. Fluorogen-activating proteins: beyond classical fluorescent proteins. Acta Pharm. Sin. B 8, 339–348 (2018).
Uchinomiya, S., Ojida, A. & Hamachi, I. Peptide tag/probe pairs based on the coordination chemistry for protein labeling. Inorg. Chem. 53, 1816–1823 (2014).
Gatterdam, K., Joest, E. F., Gatterdam, V. & Tampé, R. Scaffold design of trivalent chelator heads dictates high-affinity and stable His-tagged protein labeling in vitro and in cellulo. Angew. Chem. Int. Ed. 57, 12395–12399 (2018).
You, C. & Piehler, J. Multivalent chelators for spatially and temporally controlled protein functionalization. Anal. Bioanal. Chem. 406, 3345–3357 (2014).
Bouhedda, F., Autour, A. & Ryckelynck, M. Light-up RNA aptamers and their cognate fluorogens: from their development to their applications. Int. J. Mol. Sci. 19, 44 (2018).
Zhang, H. et al. Assembling DNA through affinity binding to achieve ultrasensitive protein detection. Angew. Chem. Int. Ed. 52, 10698–10705 (2013).
Rodell, C. B., Mealy, J. E. & Burdick, J. A. Supramolecular guest−host interactions for the preparation of biomedical materials. Bioconjug. Chem. 26, 2279–2289 (2015).
Wang, L., Li, L.-L., Fan, Y.-s & Wang, H. Host–guest supramolecular nanosystems for cancer diagnostics and therapeutics. Adv. Mater. 25, (3888–3898 (2013).
Liu, W., Samanta, S. K., Smith, B. D. & Isaacs, L. Synthetic mimics of biotin/(strept)avidin. Chem. Soc. Rev. 46, 2391–2403 (2017).
Peck, E. M. et al. Pre-assembly of near-infrared fluorescent multivalent molecular probes for biological imaging. Bioconjug. Chem. 27, 1400–1410 (2016).
Shaw, S. et al. Non-covalently pre-assembled high-performance near-infrared fluorescent molecular probes for cancer imaging. Chem. Eur. J. 24, 13821–13829 (2018).
van Dun, S., Ottmann, C., Milroy, L.-G. & Brunsveld, L. Supramolecular chemistry targeting proteins. J. Am. Chem. Soc. 139, 13960–13968 (2017).
Webber, M. J. et al. Supramolecular PEGylation of biopharmaceuticals. Proc. Natl Acad. Sci. USA 113, 14189–14194 (2016).
Patra, M., Zarschler, K., Pietzsch, H.-J., Stephan, H. & Gasser, G. New insights into the pretargeting approach to image and treat tumours. Chem. Soc. Rev. 45, 6415–6431 (2016).
Kim, T. H. et al. Mix to validate: a facile, reversible pegylation for fast screening of potential therapeutic proteins in vivo. Agnew. Chem. Int. Ed. 52, 6880–6884 (2013).
Liu, X., Sun, J. & Gao, W. Site-selective protein modification with polymers for advanced biomedical applications. Biomaterials 178, 413–434 (2018).
Mantooth, S. M., Munoz-Robles, B. G. & Webber, M. J. Dynamic hydrogels from host–guest supramolecular interactions. Macromol. Biosci. 19, 1800281 (2019).
Liu, Y. & Hsu, S.-h. Synthesis and biomedical applications of self-healing hydrogels. Front. Chem. 6, 449 (2018).
Diba, M. et al. Self-healing biomaterials: from molecular concepts to clinical applications. Adv. Mater. Interfaces 5, 1800118 (2018).
Sun, C. et al. Polymeric nanomedicine with “Lego” surface allowing modular functionalization and drug encapsulation. ACS Appl. Mater. Interfaces 10, 25090–25098 (2018).
Kim, S. et al. Cucurbit[6]uril-based polymer nanocapsules as a non-covalent and modular bioimaging platform for multimodal: in vivo imaging. Mater. Horiz. 4, 450–455 (2017).
Zhang, S. et al. Precise supramolecular control of surface coverage densities on polymer micro- and nanoparticles. Chem. Sci. 9, 8575–8581 (2018).
Zhou, Z., Han, Z. & Lu, Z. R. A targeted nanoglobular contrast agent from host-guest self-assembly for MR cancer molecular imaging. Biomaterials 85, 168–179 (2016).
Li, Q.-L. et al. Supramolecular nanosystem based on pillararene-capped CuS nanoparticles for targeted chemo-photothermal therapy. ACS Appl. Mater. Interfaces 10, 29314–29324 (2018).
Wang, Y.-X., Zhang, Y.-M., Wang, Y.-L. & Liu, Y. Multifunctional vehicle of amphiphilic calix[4]arene mediated by liposome. Chem. Mater. 27, 2848–2854 (2015).
Hou, C., Huang, Z., Fang, Y. & Liu, J. Construction of protein assemblies by host–guest interactions with cucurbiturils. Org. Biomol. Chem. 15, 4272–4281 (2017).
Liu, Y., Yang, H., Wang, Z. & Zhang, X. Cucurbit[8]uril-based supramolecular polymers. Chem. Asian J. 8, 1626–1632 (2013).
Samanta, S. K., Moncelet, D., Briken, V. & Isaacs, L. Metal–organic polyhedron capped with cucurbit[8]uril delivers doxorubicin to cancer cells. J. Am. Chem. Soc. 138, 14488–14496 (2016).
Park, K. M., Murray, J. & Kim, K. Ultrastable artificial binding pairs as a supramolecular latching system: a next generation chemical tool for proteomics. Acc. Chem. Res. 50, 644–646 (2017).
Zhu, H. et al. Pillararene-based host–guest recognition facilitated magnetic separation and enrichment of cell membrane proteins. Mater. Chem. Front. 2, 1475–1480 (2018).
Finbloom, J. A. & Francis, M. B. Supramolecular strategies for protein immobilization and modification. Curr. Opin. Chem. Biol. 46, 91–98 (2018).
Li, G.-P., Zhang, H., Zhu, C.-M., Zhang, J. & Jiang, X.-F. Avidin–biotin system pretargeting radioimmunoimaging and radioimmunotherapy and its application in mouse model of human colon carcinoma. World J. Gastroenterol. 11, 6288–6294 (2005).
Schubert, M. et al. Novel tumor pretargeting system based on complementary l-configured oligonucleotides. Bioconjug. Chem. 28, 1176–1188 (2017).
Strebl, M. G., Yang, J., Isaacs, L. & Hooker, J. M. Adamantane/cucurbituril: a potential pretargeted imaging strategy in immuno-PET. Mol. Imaging 17, 1536012118799838 (2018).
Spa, S. J. et al. A supramolecular approach for liver radioembolization. Theranostics 8, 2377–2386 (2018).
Kim, K. L. et al. Supramolecular latching system based on ultrastable synthetic binding pairs as versatile tools for protein imaging. Nat. Commun. 9, 1712 (2018).
Sasmal, R. et al. Synthetic host–guest assembly in cells and tissues: fast, stable, and selective bioorthogonal imaging via molecular recognition. Anal. Chem. 90, 11305–11314 (2018).
Rood, M. T. M. et al. Obtaining control of cell surface functionalizations via pre-targeting and supramolecular host guest interactions. Sci. Rep. 7, 39908 (2017).
Liu, S. et al. The cucurbit[n]uril family: prime components for self-sorting systems. J. Am. Chem. Soc. 127, 15959–15967 (2005).
Welling, M. M. et al. In vivo stability of supramolecular host–guest complexes monitored by dual-isotope multiplexing in a pre-targeting model of experimental liver radioembolization. J. Control. Release 293, 126–134 (2019).
Samanta, S. K., Moncelet, D., Vinciguerra, B., Briken, V. & Isaacs, L. Metal organic polyhedra: a click-and-clack approach toward targeted delivery. Helv. Chim. Acta 101, e1800057 (2018).
Bak, M., Jølck, R. I., Eliasen, R. & Andresen, T. L. Affinity induced surface functionalization of liposomes using Cu-free click chemistry. Bioconjug. Chem. 27, 1673–1680 (2016).
Robinson, P. V., de Almeida-Escobedo, G., de Groot, A. E., McKechnie, J. L. & Bertozzi, C. R. Live-cell labeling of specific protein glycoforms by proximity-enhanced bioorthogonal ligation. J. Am. Chem. Soc. 137, 10452–10455 (2015).
Long, M. J. C., Poganik, J. R. & Aye, Y. On-demand targeting: investigating biology with proximity-directed chemistry. J. Am. Chem. Soc. 138, 3610–3622 (2016).
Cañeque, T., Müller, S. & Rodriguez, R. Visualizing biologically active small molecules in cells using click chemistry. Nat. Rev. Chem. 2, 202–215 (2018).
Xi, W., Scott, T. F., Kloxin, C. J. & Bowman, C. N. Click chemistry in materials science. Adv. Funct. Mater. 24, 2572–2590 (2014).
Leppiniemi, J. et al. Bifunctional avidin with covalently modifiable ligand binding site. PLOS ONE 6, e16576 (2011).
Saunders, M. J. et al. Fluorogen activating proteins in flow cytometry for the study of surface molecules and receptors. Methods 57, 308–317 (2012).
Ouellet, J. RNA fluorescence with light-up aptamers. Front. Chem. 4, 29 (2016).
Zhang, J. X. et al. Predicting DNA hybridization kinetics from sequence. Nat. Chem. 10, 91–98 (2018).
Assaf, K. I. & Nau, W. M. Cucurbiturils: from synthesis to high-affinity binding and catalysis. Chem. Soc. Rev. 44, 394–418 (2015).
Murray, J., Kim, K., Ogoshi, T., Yao, W. & Gibb, B. C. The aqueous supramolecular chemistry of cucurbit[n]urils, pillar[n]arenes and deep-cavity cavitands. Chem. Soc. Rev. 46, 2479–2496 (2017).
Liu, W., Peck, E. M., Hendzel, K. D. & Smith, B. D. Sensitive structural control of macrocycle threading by a fluorescent squaraine dye flanked by polymer chains. Org. Lett. 17, 5268–5271 (2015).
Gómez-Durán, C. F. A., Liu, W., Betancourt-Mendiola, M. L. & Smith, B. D. Structural control of kinetics for macrocycle threading by fluorescent squaraine dye in water. J. Org. Chem. 82, 8334–8341 (2017).
Ogoshi, T., Yamagishi, T.-a. & Nakamoto, Y. Pillar-shaped macrocyclic hosts pillar[n]arenes: new key players for supramolecular chemistry. Chem. Rev. 116, 7937–8002 (2016).
Sadrerafi, K., Moore, E. E. & Lee, M. W. Association constant of β-cyclodextrin with carboranes, adamantane, and their derivatives using displacement binding technique. J. Incl. Phenom. Macrocycl. Chem. 83, 159–166 (2015).
Acknowledgements
The authors are grateful for a grant from the US National Institutes of Health (GM059078) and AD&T Berry Family Foundation fellowship from the University of Notre Dame.
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Schreiber, C.L., Smith, B.D. Molecular conjugation using non-covalent click chemistry. Nat Rev Chem 3, 393–400 (2019). https://doi.org/10.1038/s41570-019-0095-1
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41570-019-0095-1
- Springer Nature Limited
This article is cited by
-
Adaptive insertion of a hydrophobic anchor into a poly(ethylene glycol) host for programmable surface functionalization
Nature Chemistry (2023)
-
Cucurbituril curiosities
Nature Chemistry (2023)
-
The 2022 Nobel Prize in Chemistry for the development of click chemistry and bioorthogonal chemistry
Analytical and Bioanalytical Chemistry (2023)
-
Hydrogels for Cardiac Restorative Support: Relevance of Gelation Mechanisms for Prospective Clinical Use
Current Heart Failure Reports (2023)
-
Switchable bifunctional molecular recognition in water using a pH-responsive Endo-functionalized cavity
Nature Communications (2022)