Abstract
Computer-aided design of novel molecules and compounds is a challenging task that can be addressed with quantum computing (QC) owing to its notable advances in optimization and machine learning. Here, we use QC-assisted learning and optimization techniques implemented with near-term QC devices for molecular property prediction and generation tasks. The proposed probabilistic energy-based deep learning model trained in a generative manner facilitated by QC yields robust latent representations of molecules, while the proposed data-driven QC-based optimization framework performs guided navigation of the target chemical space by exploiting the structure–property relationships captured by the energy-based model. We demonstrate the viability of the proposed molecular design approach by generating several molecular candidates that satisfy specific property target requirements. The proposed QC-based methods exhibit an improved predictive performance while efficiently generating novel molecules that accurately fulfill target conditions and exemplify the potential of QC for automated molecular design, thus accentuating its utility.
Similar content being viewed by others
Introduction
The development of novel compounds and materials has played a critical role in the advancements of various scientific fields. Technological and societal progress can be further fueled by the discovery of novel molecules for applications ranging from drug design for treating diseases to efficient energy storage devices for combating climate issues1. The potential for the synthesis of novel molecular compounds remains vast; for instance, the number of molecules with pharmacological properties that could be synthesized is estimated to be as high as 1060 2. Computer-aided molecular design can facilitate the generation of molecular candidates with desired chemical properties through various means3. Simulations of chemical systems could estimate molecular properties through quantum chemistry calculations. However, these techniques are unable to handle large-scale systems through first-principle calculations and require the use of approximations adopted at the expense of accuracy, which may inhibit efficient exploration of the chemical space4. Computational optimization techniques have also demonstrated some success in the generation of useful molecules5. Additionally, machine learning, specifically deep learning, has accelerated progress in molecular design through learning patterns in molecular datasets and offers a promising route toward the development of novel compounds6. The complexities stemming from the nonlinearity of molecular design problems at larger scales lead to the intractability of optimization techniques7, while the machine learning techniques may not be data-efficient and yield inaccurate predictions despite consuming prohibitively large computational power to process huge amounts of molecular data. It is imperative to exploit efficient computational techniques for a guided exploration of the chemical space to generate insights into the synthesis of novel molecules.
Reliable techniques for molecular property prediction and efficient search strategies are the building blocks for computer-aided molecular design8. Prediction models that can estimate the properties of given molecules can assist the virtual screening process in isolating candidate molecules with the desired properties9. Computational screening of molecules is dependent on the quality of virtual chemical libraries manually constructed from chemical databases10 or through combinatorial approaches11,12, and may induce uncertainty in the exploration of the appropriate chemical space. In addition to using first-principle simulations for predicting molecular properties13, machine-learning techniques have also led to the development of advanced prediction models that estimate properties with competitive precision as that of traditional techniques14. Such data-driven methods circumvent the need for computationally expensive quantum chemical methods15 and have been more commonly embraced to improve the performance of predictive models16. Characterization of molecular structure–property relationships through interpretation of machine learning models can be further used to guide the design of novel molecules and is referred to as inverse molecular design17. Inverse design can be performed by navigating the chemical space of molecules with target functionality through optimization, search, or sampling techniques18. Several optimization techniques, including both heuristic and deterministic algorithms, can be applied to inverse molecular design cast as an optimization problem. Evolutionary techniques like genetic algorithms19 and discrete combinatorial optimization approaches like mixed-integer programming20 have demonstrated their utility for the design of various molecules. However, genetic algorithms require manual adjustment of heuristic rules for different optimization problems and do not guarantee optimality, while combinatorial optimization approaches may exhibit difficulty in solving large-scale nonlinear optimization problems21. These computational challenges can be tackled by deep learning methods that utilize sophisticated neural network architectures for constructing generative models for molecular design. Various deep generative models allow for adopting different molecular representations as input and can be trained to learn the distribution of the molecular dataset6, which is followed by random sampling of molecules for further screening with property estimation models. Recurrent neural networks22,23 and variational autoencoders24,25 that use SMILES identifier as input, and graph convolutional neural networks (GraphConv)26 that operate on molecular structures represented by graphs are some of the commonly used deep learning architectures for molecular design. Deep learning approaches can also be tailored for the conditional generation of molecules that satisfy the target property requirements27. In addition to direct sampling from trained generative models, Bayesian optimization24 and reinforcement learning28, assisted by neural network architectures for property estimation, can perform a guided search for molecules through additional optimization steps often carried out in the latent space of the neural networks. Despite their potential, the dependence of deep neural networks on large amounts of diverse training data comprising molecules with properties spanning the chemical space may cast doubt on their generative abilities in the presence of out-of-domain uncertainty.
Quantum computing (QC) holds tremendous potential to achieve significant technological feats in various domains, including the design of novel molecules for specific purposes29. Owing to their ability of exploiting quantum mechanical phenomena for performing computation, QC techniques have demonstrated remarkable improvements for several applications. The promise of performance enhancement offered by QC has also attracted considerable attention from the research community for the development of QC-based methods in fields like computational chemistry, optimization, and machine learning30. Quantum computers offer a fundamentally different approach to performing quantum chemistry simulations that have helped overcome the practical challenges of simulating chemical systems on classical computers31. Quantum algorithms have also facilitated the development of quantum-enhanced optimization and machine learning techniques tailored for specific problem types and learning tasks32,33. Recently, QC algorithms have also been proposed for the search of optimal configuration of molecules in protein chains that demonstrate a quantum speedup over straightforward enumeration34. Despite their advantages, QC techniques implemented on current quantum devices exhibit limitations in terms of performance and scalability due to the presence of hardware noise and a limited number of quantum bits or qubits35. Therefore, it is important to develop models and methods that harness the complementary strengths of high-performance quantum and classical computation by overcoming their individual limitations in order to effectively and efficiently navigate through the complex chemical space for molecular design. Although quantum-enhanced machine learning and optimization can be employed for molecular property prediction and inverse design, several research challenges remain. Developing prediction models and design methods that are compatible with near-term quantum devices with noisy qubits is the first challenge. There have been attempts at hybrid quantum-classical optimization techniques for determining the structural configuration of molecules36,37, but these approaches do not scale for larger molecules on today’s quantum computers. As a result, scalable QC approaches for a molecular design that can handle problems across varying scales are another important research challenge. An additional one lies in exploiting the noisy nature of near-term intermediate-scale quantum (NISQ) devices which comprise 50 to a few hundred qubits without fault-tolerant capabilities35 for learning and inverting structure–property relationships without compromising performance.
In this paper, we propose a hybrid quantum-classical computational framework for molecular design that utilizes QC-based learning and optimization strategies to efficiently navigate the chemical space for targeted molecular generation. We construct an energy-based deep learning model that can be trained with QC-assisted generative training to extract latent representations from molecular graphs for property prediction. A QC-assisted optimization technique is further presented that efficiently traverses through the chemical space to identify molecules with desired properties by inverting the structure–activity relationship captured with the energy-based model. These are particularly advantageous when exploring the chemical space for candidate molecules by not relying solely on the available molecular datasets. A thorough analysis of the proposed methods that includes benchmarking against the baseline models and investigating the efficacy of the molecular generation pipeline is also conducted. The proposed model learns the conditional distribution of molecular properties depending on their structural information allowing us to efficiently predict properties for a given molecule. Consequently, it learns the relationship between molecular composition and structure and its physiochemical properties, which is exploited by the proposed optimization technique to generate molecules exhibiting target properties. Previous deep learning methods for molecular generation do not always facilitate constrained sampling of molecular candidates6. In contrast, our molecular design framework is designed to generate molecules given a certain composition that exhibit target property values within specific ranges. Upon benchmarking with existing deep learning-based approaches, the proposed molecular design framework not only demonstrates competitive predictive performance for physiochemical properties but also efficiently generates molecules for predefined property requirements. The computational experiments conducted to validate the generated molecules and their properties obtained with various molecular design approaches also reveal the data efficiency and generalization capabilities of the proposed QC-based molecular design approach through the exploration of sparsely populated regions of the chemical space.
The major contributions of this study are summarized as follows:
-
A data-efficient hybrid quantum-classical approach for molecular property estimation leverages a deep learning model trained with a QC-assisted learning approach to extract robust latent representations of molecules.
-
A QC-based approximate optimization technique exploiting the trained property estimation model to explore the chemical space in a guided manner and identify molecular candidates with desired properties.
-
Several druglike molecules are generated with the proposed QC-based molecular design framework for various physiochemical property targets in an efficient manner as compared to existing deep learning-based molecular design approaches.
Results and discussion
QC-assisted molecule generation framework
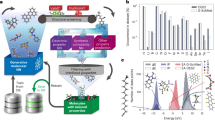
This study utilized quantum annealing-based strategies for learning and optimization required for molecular generation. We first construct an energy-based model to learn the distribution of molecular properties conditioned on corresponding fingerprints. A GraphConv network with fixed weights is employed to generate fixed-length neural fingerprints, as illustrated in Fig. 1a. The only input to this model is the structural information of the molecule describing the atom types and their connectivity26. The constructed energy-based model uses the generated molecular descriptors f and the molecular property range y as the input data. This energy-based model is trained with a set of molecule–property pairs by drawing samples from a quantum annealer to estimate the gradients required for parameter update rules. Upon training, the constructed energy-based model learns the probability distribution \(p(y|f)\), as shown in Fig. 1b. The conditional energy-based model also utilizes latent variable representations h that can be considered as the compressed chemical space spanned by the molecules and their properties. These latent representations can be further used to perform molecular property estimation tasks by passing them as input to a separate feedforward network. For a molecular generation, we employ an iterative optimization procedure that utilizes a quantum annealer to solve formulated quadratic unconstrained binary optimization (QUBO) problems. As illustrated in Fig. 1c, a surrogate model is constructed to estimate the free energy of the molecule–property pair with the trained conditional energy-based model. After formulating a QUBO problem that integrates the linear surrogate model with structural constraints, the problem is then solved using a quantum annealer to generate potential molecular candidates. Governed by the proposed optimization procedure, the surrogate model is sequentially refined to explore the chemical space for identifying molecules that satisfy the desired property requirements and structural constraints.
The energy-based model is trained by drawing samples from a quantum annealer in (b) and captures the structure–property relationship between molecular representations or descriptors generated with a GraphConv network in (a) and the molecular properties. The trained conditional energy-based model is used to estimate the free energy of input molecules and compute objective values in (c). Formulating and solving quadratic unconstrained binary optimization problems in an iterative manner with a quantum annealer in (c) yields molecular design candidates with desired target properties.
Molecular property prediction
Constructing an efficient molecular property prediction model that can provide insights into the structure–property relationships is an important first step toward guiding the generation of molecules with desired properties. The latent representations of the molecules generated by the proposed conditional energy-based model play an important role in predicting molecular properties. The predictive performance of feedforward models that use various inputs obtained through different methods is presented in Table 1. The average and standard deviation values of the mean absolute error computed over multiple training-evaluation repetitions are reported here. For the predictive models that utilize the latent representations of the energy-based model as inputs, we obtain several sets of these representations by training multiple conditional energy-based models with both CD learning and QC-assisted learning. With each latent representation obtained with the corresponding energy-based model, repeated experiments for property prediction are performed with feedforward networks to measure the relevant metrics along with their statistical measures. Among the baseline predictive models that use rule-based molecular descriptors as input, the larger ECFP fingerprint tends to perform better for predicting QED and LogP, while the predictive model with MACCS yields much more accurate predictions for the accessibility scores. The baseline model, which uses neural fingerprints generated by the GraphConv model, produces significantly higher errors when predicting the drug-likeness property of the molecule but obtains competitive results for the remaining molecular properties with only a 13.6% increase over the lowest error associated with the synthetic accessibility scores. On the other hand, the latent variable representations obtained with the conditional energy-based models trained with both classical and QC-assisted learning techniques achieve an accurate predictive performance for all property targets despite their low dimensionality. The predictive models that use latent representations obtained with QC-assisted generative training not only yield competitive prediction errors as that of other baseline models but also enjoy the least observed error when predicting the drug-likeness of the molecules. The obtained computational results demonstrate the effectiveness of the latent representations obtained with the conditional energy-based model trained with QC-assisted learning for molecular property prediction. We also generate the latent representations of 1000 molecules in the test set with their known target labels using the conditional energy-based model trained through QC-assisted generative training. The mapping of these representations along their three principal components is plotted in Supplementary Fig. 1 for various molecular properties.
Targeted molecule generation
We utilize the trained energy-based model and impose restrictions on the molecular properties for a targeted molecular generation with the proposed QC-assisted optimization technique. Several target conditions are imposed on the drug-likeness of molecules as well as their partition coefficients. Table 2 presents the generation statistics for the molecules generated with the QC-assisted optimization technique along with their computed properties. This table also comprises identical statistics for the molecules in the training set that satisfy the corresponding target requirements. For the chosen property targets, we benchmark the selected baselines and report the corresponding properties of the molecules obtained with these molecular design techniques. The number of generated molecules obtained by exploring the chemical space described by each property target range and the atomic identities of the reference molecules in the test set are reported in this table. Among the generated molecules for a given target condition, a few duplicates are also identified that form <1.5% of the overall molecules. During the sampling of molecules performed by the QC-assisted approximate optimization technique, the validity of the molecules is verified with the RDKit package38. Although invalid molecules that fail to comply with structural constraints are rarely detected owing to the feasible molecular generation capability of the QC-assisted approach, they are automatically discarded to prevent inaccurate estimations computed within this data-driven approach. Furthermore, the average and standard deviation values presented for each target condition are calculated over the observed properties of the generated molecules to ensure the reliability of the molecular generation process subject to conditions. As evident from the computational results in Table 2, the molecules generated with the QC-based approach for each conditioned property range satisfy the respective imposed restrictions. For some property targets, the deep learning approaches CVAE and MGM are able to generate molecules that comply with the requirements. On the other hand, the genetic algorithm GBGA is unable to achieve efficient targeted molecular generation and may require manual adjustment of the fitness function for each property target. We also plot the distribution of the SAS scores for all generated molecules and different target properties in Fig. 3c, d to generate insights for their ease of synthesis. From these violin plots, it can be seen that the molecules generated with lower QED and LogP values exhibit wider variability of accessibility scores, despite their relatively higher average SAS scores. A reverse trend is observed for molecules generated for higher utility as drug-like molecules and high LogP values, demonstrating lower variability of SAS scores with low average scores. We also investigate the latent representations for the generated molecules obtained with the trained conditional energy-based model using the t-SNE embeddings. Mapping these latent representations in 2D allows us to identify the closeness of molecules with respect to their corresponding properties. Figure 2 shows the 2D embeddings obtained with t-SNE for the molecules in the training set as well as the generated molecules colored according to the QED property values. We also include a few examples of molecular structures obtained for different property targets in this figure. A distinction between the latent representations for various property ranges can be observed with molecules exhibiting similar QED values adjacent to each other. This illustrates that the constructed energy based-model learns to capture the relationship between the molecules and their properties as molecules with similar properties have close embeddings.
To assess the effects of data used to capture the structure-property relationship on the exploration of the chemical space, the densities of partition coefficients and QED values are visualized with kernel density estimation (KDE) plots along with their marginal distributions for the molecules in the training set and all generated molecules in Fig. 3a, b. The observed concentration of molecules in both training and generated sets is highest in approximately similar ranges of molecular properties. Figure 3a, b shows that the molecules generated exhibit higher density levels when they have either low partition coefficients and high QED values or high partition coefficients and lower QED values. However, this trend is missing for the samples in the training set. In addition to LogP and QED, we also compute the Kullback–Leibler (KL) divergence values for various molecular properties to measure the difference between the distribution of generated molecules with that of the training set distributions. The KL-divergence scores for the molecules generated with the proposed QC-based framework, along with the CVAE, MGM, and GBGA baselines, are reported in Supplementary Table 5. With the exceptions of the number of hydrogen bond acceptors and internal similarity, the molecules generated with the QC-based molecular design approach exhibit the highest KL-divergence scores as compared to the other baselines. These results indicate that, unlike deep generative models that sample molecules from the distribution captured from the training set, the proposed QC-based optimization technique is able to efficiently traverse through the unexplored chemical space that is not previously mapped with the structure–property relationship described by the training set samples. Examples of generated molecular structures sampled for various atomic identities and target properties are also provided in Fig. 3e, f. Although the proposed approximate technique for molecular generation requires extra steps as compared to direct sampling from deep generative models, the QC-assisted technique yields a diverse set of feasible molecules that satisfy structural constraints through efficient guided optimization performed within the target chemical space.
The KDE plots indicate the density of partition coefficient (LogP) and quantitative estimation of drug-likeness (QED) for the a molecules in the training set and b the generated molecules. The distribution of synthetic accessibility scores (SAS) for the generated molecules is visualized with violin plots for target conditions on c QED and d LogP, respectively. Examples of generated molecular structures conditioned upon restrictions on molecular properties of e QED and f LogP are also provided.
Validation
The validity of the generated molecules obtained with different molecular design frameworks is verified along with their properties, followed by plotting the distributions of the properties for corresponding property targets in Fig. 4. Distributions for the training data are also plotted to validate the generalization capabilities of the molecular design techniques. As evident from these density plots, the CVAE architecture does not guarantee constrained sampling of molecular candidates. The graph-based deep learning model MGM is able to generate a few molecules that satisfy corresponding targets but is accompanied by the generation of other noncomplying structures in a significant proportion. As evident from the efficacy metrics reported in Table 2, a deviation as high as 72.3% from the mean target property requirements can be observed with the baseline deep learning methods for molecular generation. In contrast to the graph and autoencoder-based baselines, GBGA limits the search of molecules candidates within a narrow domain for the QED property targets but exhibits wider exploration for the LogP property targets. On the other hand, the proposed QC-based approach is able to generate molecules that exhibit target properties efficiently with observed zero violations of the required target property constraints. Additionally, the generated molecules with the proposed molecular design technique follow trends similar to that of the training data, which evidently demonstrates the learning and data efficiency of the proposed energy-based model trained with QC-assisted learning. Owing to the reliability of the QC-based molecular design framework in terms of accurate property prediction and efficient targeted molecular design, the proposed strategies can be easily adopted in laboratories for experimental validation. The efficacy of the presented QC-based techniques implemented on noisy near-term quantum devices like quantum annealers has further illustrated the promise of QC for the design of novel molecules in the NISQ as well as the fault-tolerant era.
Distributions of the molecular properties of molecules generated with various molecular design frameworks, including the proposed QC-based technique, conditional variational autoencoder (CVAE), masked graph model (MGM), and graph-based genetic algorithm (GBGA). The molecules are generated with these frameworks for different property targets for QED (a–e) and LogP (f–j), as shown in the figure. The distribution of the properties for molecules in the training set satisfying the corresponding targets is also provided for reference.
The computational results presented in the above subsections have demonstrated the applicability of QC-assisted techniques for molecular property estimation and design. Although the goal of this work is to develop a molecular design framework capable of exploiting QC techniques for efficient molecular property estimation and generation, it is essential to justify the use of QC for learning and optimization tasks. To this end, we compare the performance of the proposed QC-based techniques with that of their classical counterparts. Computational experiments conducted here use quantum annealers for training the conditional energy-based model as well as for sampling molecules within the proposed optimization technique. As seen in Table 1, the predictive performance of the model that uses latent representations generated through QC-assisted training is significantly better than the baseline predictive models but exhibits only slight improvement over latent variables obtained with classical CD-learning. To demonstrate the training efficiency of the energy-based model with QC-assisted generative training, the training curves obtained over multiple runs are plotted in Fig. 5a. As evident from these, QC-assisted generative training progresses at a rate similar to that of CD learning but converges to a significantly lower free energy difference value. Additionally, by drawing samples directly from the quantum annealer, QC-assisted learning requires \(O(|y|\cdot |h|\cdot (|y|+|h|))\) less algebraic operations than one-step CD learning for each step of the training process, where y and h represent the one-hot encoded vector denoting the property target label and the latent vector, respectively. The quality of training in energy-based models directly impacts the molecular design capabilities of the QC-assisted optimization technique. This is validated by utilizing two conditional energy-based models trained with CD learning as well as QC-assisted learning for estimating and minimizing associated free energy. We fix the reference molecules used as the initial basis for the optimization procedure along with the number of exploration and optimization steps and measure the number of molecules obtained with each conditional energy-based model that satisfies the various property requirements. Distributions of the proportion of the total molecules generated with both conditional energy-based models are presented in Fig. 5c, d for the QED and LogP property targets. It can be observed that for most targets, the number of identified molecules with desired properties is higher for the energy-based model trained with QC-assisted learning than one with CD learning. To verify whether the conditional distribution captured by the energy-based model plays a role in locating the desired molecular candidates, we plot the optimization trajectories for identical reference molecules of varying sizes used with both models in Supplementary Fig. 2. As evident from the trajectory curves for CD learning, the approximate optimization technique used with the conditional energy-based model is subject to higher levels of stochasticity than its quantum counterpart, which yields molecules in a structured manner. As evident from the performance metrics associated with these optimization trajectories in Supplementary Table 3, the proposed optimization technique used with an energy-based model trained with QC-assisted learning exhibits improved performance in terms of speed of improvement as well as best-found solutions. The proportion of molecules obtained with the classically trained energy-based model in Fig. 5c, d can also be attributed to this random exploration of the chemical space. Figure 5b also presents a comparison of computational times required to solve the QUBO problems of varying sizes that occurred during molecular generation for different atomic compositions and property targets with both simulated annealings implemented on a classical computer and quantum annealing. In addition to the generation efficacy of the proposed QC-based molecular design, we also evaluate the computational resource utilization associated with solving QUBO problems in the approximate optimization technique. QUBOs constructed for reference molecules of varying sizes are collected and solved with both simulated annealing and quantum annealing with the graph and time metrics reported in Supplementary Table 4. The 50th, 75th, and 90th percentiles of the annealing times are also plotted in Fig. 5b. The computational time required to solve QUBO problems arising during the optimization procedure for molecular design increases with the size of the reference molecules. On the other hand, the solution time for the quantum annealer does not vary with the problem size and is even approximately 200 times faster for the largest problem instance solved with simulated annealing implemented on a classical computer. The presented comparisons clearly demonstrate that the QC-assisted techniques serve as an enhanced alternative to conventional learning and optimization methods implemented on classical computers.
a Learning curve for the conditional energy-based model trained with QC-assisted generative training and CD learning, b the 50th, 75th, and 90th percentiles of annealing times over a set of 25 instances for both simulated and quantum annealing. The distribution of the proportion of molecular candidates satisfying target requirements obtained with the energy-based models trained with both CD learning and QC-assisted learning are plotted for c QED property targets and d LogP property targets. The same set of reference molecules is used as the initial starting point for optimizing molecules with both models for a fair comparison.
Computational issues
One of the limitations of the proposed QC-assisted strategy for molecular generation is that the exploration of molecular candidates exhibiting target properties can only be performed within certain ranges only. Although this constrained exploration of the chemical space provides better flexibility in identifying molecules with known atomic identities that satisfy the property requirements, determining molecules with exactly given properties may be difficult. In contrast to the trained deep generative models that can sample molecules for specific target properties, we apply an optimization procedure to perform a guided search for molecules that satisfy the target requirements. This is evident from the presented properties of generated molecules obtained with various molecular design techniques in Table 2 and Supplementary Table 3. Within our computational experiments, we observed that the molecules that exhibit desired properties were found generally towards the end of the optimization procedure, which requires a higher computational effort than direct sampling with deep generative models. We acknowledge that the proposed QC-based strategies for molecular design exhibit certain limitations as compared to the generative deep learning systems for molecular generation in literature. Despite these limitations and the challenges associated with near-term QC, comparisons of property prediction with various baselines followed by an analysis of the generated molecules substantiates our objective of developing a QC-based molecular design framework capable of generating molecules with high efficacy. Areas for future improvements involve the development of generative models that could be trained efficiently with QC-based strategies and are capable of directly sampling molecular candidates without any additional steps. Furthermore, the use of the presented QC-based strategies to study and generate chemical reactions could also be explored.
Methods
Data
We use compounds from the Zinc database39 to train and validate the performance of the proposed methods. A subset of Zinc comprising 12,000 molecules that are commonly used for benchmarking purposes is collected for our computational study40. The collected SMILES identifiers of the molecules are converted to graph-structured data by identifying the node features and edge features using the RDKit package38. The node features distinguish constituent atoms into nine heavy atom types, while the edge features describe the presence of bonds between atoms and bond types. For each molecular graph in the dataset, we also collected three different properties, namely, the quantitative estimation of drug-likeness (QED)41, Wildman–Crippen partition coefficient (LogP)42, and the synthetic accessibility score (SAS)43. The LogP value or the water-octanol partition coefficient provides a measure of lipophilicity and serves as one of the molecular properties used to estimate QED. On the other hand, the SAS values describe the ease of synthesis of drug-like molecules. The QED property can vary between zero and one, with a larger value indicating a more drug-like molecule while the SAS ranges from one to ten. Among the molecules in the selected dataset, LogP ranges from -4.58 to 6.66. The averages of QED, SAS, and LogP for the collected samples are 0.728, 3.048, and 2.450, and the standard deviations for these are 0.138, 0.834, and 1.433, respectively. Target labels for each molecule and its corresponding property are also generated by discretizing the target space into several bins. Bin ranges for each property are provided in Supplementary Table 1.
Molecular representation
The first step towards constructing quantitative structure-property relationships is to transform the chemical information of molecules into descriptors or fingerprints. The numerical encoding of the molecular structures is necessary for their feature representation as well as for similarity searches during virtual screening. MACCS and ECFP are examples of substructure-based and circular fingerprints, respectively44. Although such fingerprints have been previously used for predictive modeling, they are expert-based representations that are constructed with expert knowledge governed by a set of rules45. In addition, the non-invertibility of molecular fingerprints further complicates the search for molecules within the chemical space that produce desired representations obtained with predictive models. As a result, we use a graph convolutional neural network to generate neural fingerprints that serve as input to the energy-based model. Graph convolutional networks allow for learning task-specific molecular features to achieve a competitive predictive performance and are capable of generating molecular fingerprints analogous to circular fingerprints like ECFP46. To generate neural fingerprints, a graph convolutional neural network is constructed that operates on molecules structured as graphs with atoms as nodes and bonds as edges. We fix the weights of the graph neural network to generate a direct non-adaptive mapping between the molecular structure and the generated fingerprint and exploit their versatility. The constructed network uses a convolution operator described in46 followed by a linear transformation to generate hidden feature vectors for individual atoms. This convolution operator consists of distinct weight matrices for each possible degree of vertices in the input graph. A softmax operation along the node features is then performed for each node to facilitate information flow between the atomic features of the molecule. Finally, a global pooling operation is applied to the updated hidden node features to generate a fixed-length vector as the network output. It should be noted that the graph convolutional network is initialized with large random weights without training. This graph convolutional network yields a direct nonlinear mapping Gc between the molecular graph and its neural fingerprint \(f={G}_{{\rm {c}}}(X,A)\), where X represents the feature matrix for nodes and A represents the adjacency matrix of the graph. The neural fingerprints are validated in Supplementary Note 4 through similarity comparisons with ECFP and MACCS molecular representations.
Drawing samples from quantum annealer
Quantum annealing refers to a search technique that can be implemented on an AQC platform. Compared to simulated annealing, quantum annealing demonstrates convergence to the ground state with a larger probability under similar conditions47. Quantum annealers are made available by D-Wave Systems with access to their users through the cloud. The D-Wave system can only solve quadratic unconstrained binary optimization (QUBO) problems. The QUBO graph is then mapped onto the quantum processing unit (QPU), with edges between nodes as internal couplers and qubits as the nodes. D-Wave offers QPUs with a topological structure of interconnected qubits forming either the Chimera graph or Pegasus topology48. To physically perform this mapping from variables to qubits, the process of minor embedding is required49,50. Solving a QUBO problem on the D-Wave QPU comprises three main steps. First, the QUBO problem graph is embedded in the QPU. Second, low-energy solutions to the problem Hamiltonian are obtained through quantum annealing. Third, the solution is unembedded and is returned to the user. Notably, the process of finding an optimal graph-minor embedding is NP-hard. However, heuristic algorithms exist for finding graph minors51, and the D-Wave system implements its automatic embedding and unembedding tools52. Nevertheless, special care must be taken to formulate the smallest QUBOs possible to facilitate the embedding process for large-scale problems, and to enable the solution on such quantum devices. Quantum annealers allow for the implementation of a target Ising Hamiltonian with only pairwise interactions and local terms as \({H}_{{\rm{Ising}}}={\sum }_{i,j}{J}_{ij}{\sigma }_{i}^{z}{\sigma }_{j}^{z}+{\sum }_{i}{h}_{i}{\sigma }_{i}^{z}\) and can generate independent configurations from noisy Gibbs distributions53. HIsing indicates the Ising Hamiltonian representing the quantum form of the QUBO problem, such that eigenvalues of the Ising Hamiltonian coincide with the least energy solution minimizing the corresponding QUBO. All computational experiments involving the use of quantum annealers in this work are performed with the D-Wave Advantage quantum processor that offers at least 5000 qubits and 35,000 couplers54. Each annealing run used for both generative training and optimization is performed for 20 µs on this quantum processor.
Energy-based model
We develop an energy-based model for molecular property prediction that can utilize QC techniques for efficient learning. Energy-based models can learn the distribution of data by associating an unnormalized probability value or energy to each data point. Additionally, the difficulty in sampling from such models allows us to explore alternatives for classical approximation techniques with quantum sampling facilitated by a quantum computer. In this work, we adopt a conditional generative model called conditional restricted Boltzmann machine (CRBM) to incorporate molecular property targets as binary variables. CRBM is a nonlinear generative model that can capture the conditional probability of observed data and has been previously applied for time series generation55. Although energy-based models are typically used for generative modeling, they can also be used for classification tasks56. Owing to their rich expressivity of latent variables and modeling flexibility, we use a conditional energy-based model for the supervised learning task of molecular property prediction.
An input to the energy-based model is a fixed-length vector description of the molecule under consideration captured by the neural fingerprint. In order to learn the conditional distribution \(p({\bf{y}}|{\bf{f}})\) of property targets y given the fingerprint f, we construct a CRBM network that defines a conditional joint distribution over f and latent variables h denoted by \({p}_{\theta }({\bf{y}},{\bf{h}}|{\bf{f}})\). The property targets are encoded into one-hot vectors through discrete binning of the target property values to minimize the effects of small observation errors. The hidden latent variables are modeled as binary units and can provide auxiliary information. The marginal conditional distribution of the target property y given the molecular fingerprint f can then be written as shown in Eq. (1), where \({F}_{\theta }({\bf{y}},{\bf{f}})\) represents the associated free energy and \(Z({\bf{f}})\) denotes the partition function, as defined in Supplementary Method 3. The constructed conditional energy-based model can be trained over a dataset D by maximizing the conditional log-likelihood of the input data as \({\max }_{\theta }{\sum }_{d\in D}\log {p}_{\theta }({{\bf{y}}}^{d}|{{\bf{f}}}^{d})\). In order to apply gradient ascent for this maximization problem, the gradients of conditional log-likelihood can be analytically computed which leads to the parameter update rules as derived in Supplementary Method 3. The \({\langle \cdot \rangle }_{{\rm {data}}}\) terms in the update rules can be easily computed with training data comprising of molecular fingerprints and properties for different molecules, however, computing the \({\langle \cdot \rangle }_{{p}_{\theta }({\bf{y}},{\bf{h}}|{\bf{f}})}\) exactly requires evaluating an expectation over \({p}_{\theta }({\bf{y}},{\bf{h}}|{\bf{f}})\). Estimation of this expectation value is computationally intractable due to the complexity stemming from the partition function \(Z({\bf{f}})\) and requires appropriate approximation techniques in a practical setting.
Contrastive divergence (CD) learning can approximate the intractable expectations with Gibbs sampling57 and has been extensively used for training energy-based models. CD-learning can, however, produce biased estimates of the conditional log-likelihood gradients and may even fail to converge in some cases. Although improved variants of CD learning can tackle such issues, the improvement in training performance is obtained at the expense of increased computational burden56. To overcome the limitations of classical approximation techniques, we incorporate QC-assisted training methodologies for efficient training of our energy-based model. \({p}_{\theta }({\bf{y}},{\bf{h}}|{\bf{f}})\) is a Boltzmann distribution also termed as Gibbs distribution expressed in the form of \(p(s)=\exp (-E(s))/Z\), wherein the energy E resembles an Ising Hamiltonian. As a result, we exploit quantum annealers to model \({p}_{\theta }({\bf{y}},{\bf{h}}|{\bf{f}})\) and draw samples from them to train the constructed energy-based model. The target Ising Hamiltonian on the quantum annealers is specified through the local fields h and pairwise couplings J. These input parameters are computed by mapping the energy function of the conditional energy-based model \(E({\bf{f}},{\bf{h}})=-{\sum }_{i}({b}_{i}+{\sum }_{k}{W}_{1}^{ki}{f}_{k}){y}_{i}-{\sum }_{j}({c}_{j}+{\sum }_{k}{W}_{2}^{kj}{f}_{k}){h}_{j}-{\sum }_{i,j}{W}^{ij}{y}_{i}{h}_{j}\) to the Ising Hamiltonian. This energy function can be cast as a logical bipartite graph with y and h as its nodes and the parameters \(\theta \equiv ({W}_{1},{W}_{2},W,b,c)\) and fingerprint f governing its node and edge weights. In order to draw samples from the quantum annealer, we map the bipartite graph onto the physical hardware graph of qubits and couplers. It is important to note that the connectivity of the hardware graph poses certain restrictions on the size of logical graphs that can be embedded into the QPU, so careful consideration of the degree of the logical bipartite graph dependent on the sizes of fingerprint and latent variable vectors is required. We obtain an embedding scheme for mapping the energy function onto the quantum annealer with a heuristic tool51 which is used repeatedly while drawing samples from the Boltzmann distribution. For each update step of the model parameters, several samples are drawn from \({p}_{\theta }({\bf{y}},{\bf{h}}|{\bf{f}})\) by performing multiple annealing runs to estimate the expectations \({\langle {\bf{y}}\rangle }_{{p}_{\theta }({\bf{y}},{\bf{h}}|{\bf{f}})}\), \({\langle {\bf{h}}\rangle }_{{p}_{\theta }({\bf{y}},{\bf{h}}|{\bf{f}})}\), and \({\langle {{\bf{y}}}^{{\rm {T}}}{\bf{h}}\rangle }_{{p}_{\theta }({\bf{y}},{\bf{h}}|{\bf{f}})}\).
Property estimation
The trained conditional energy-based model yields the marginal distribution \({p}_{\theta }({\bf{y}}|{\bf{f}})\) due to the generative modeling approach. For property prediction tasks, we can estimate the target property by computing the latent representations with \({p}_{\theta }({\bf{h}}|{\bf{y}},{\bf{f}})\) and training a feedforward neural network using these representations as input. For molecules with unknown targets, the latent variables can be estimated by first determining the appropriate target label y with maximization of the conditional log-likelihood given a molecular fingerprint as \({\max }_{{\bf{y}}}\,\log p({\bf{y}}|{\bf{f}})\), which is followed by computing the latent representations and passing them through the feedforward network to predict exact property targets. Fixed-length neural fingerprints of size 256 are generated with the fixed weights GraphConv network that serves as input to the conditional energy-based model in addition to the corresponding property labels encoded as discrete bins. We set the dimension of latent space representation to 64 during the construction of the energy-based model. This conditional energy-based model is trained with the molecules and their property targets in the training set by drawing samples from a quantum annealer. The same energy-based model is also trained with CD-learning57 carried out on a classical computer to discern any benefits of the QC-based approach over its classical counterpart. The training procedure of the proposed conditional energy-based models is monitored with the difference between the free energy of original and reconstructed target labels. Furthermore, three baseline models are trained to predict molecular properties for benchmarking purposes. Two fully-connected networks that use ECFP and MACCS fingerprints44 of lengths 2048 and 512 as input are trained using backpropagation with the Adam optimizer58. The same procedure is used to train a GraphConv model46 that operates directly on molecules cast as graph-structured data to predict their properties directly. The training for all models is terminated when the validation metrics fail to improve over consecutive steps, and their predictive performance is evaluated with mean absolute errors computed on the test set.
Optimization strategy for molecular design
Generation of molecules by inverting the structure-property relationships obtained with the trained conditional energy-based model can be cast as an optimization problem. For a given target property range, the corresponding one-hot label can be determined as y, then the optimization problem deals with the objective \({\min }_{X,A}{F}_{\theta }({\bf{y}},{\bf{f}})\) and is subject to the constraint \({\bf{f}}={G}_{{\rm {c}}}(X,A)\) in addition to the structural constraints \(\varOmega\) that guarantee the generation of feasible molecules. This problem can be simplified by fixing the feature matrix of atoms X in the molecule provided either by guided intuition or reference molecules, which leads to a search of the chemical space defined by the given atomic identities. Despite this, the optimization problem \({\min }_{A}\{{F}_{\theta }({\bf{y}},{\bf{f}})|{\bf{f}}={G}_{{\rm {c}}}(X,A);\,A\in \varOmega \}\) is a constrained nonlinear optimization problem with discrete-continuous variables and is challenging to solve with off-the-shelf solvers. To tackle this problem, we develop a QC-assisted approximate optimization technique that is capable of efficiently navigating the chemical space defined by the given atomic identities and the desired property targets.
In order to exploit QC for optimization, it is important to formulate problems that are compatible with the specific quantum hardware. It should be noted that both circuit model quantum devices and quantum annealers can natively solve QUBO problems. The main idea behind the proposed solution technique is the use of a weighted linear model \({\sum }_{i,j > i}{\beta }_{ij}{A}_{ij}\) that serves as a surrogate model to approximate the objective function \({F}_{\theta }({\bf{y}},{G}_{{\rm {c}}}(X,A))\). Since the adjacency matrix is symmetric, \(A\equiv \{{A}_{ij}\in \{0,1\}|j > i,\,\forall i\}\) form the variables for the molecular generation problem. In addition, the set of structural constraints \(\varOmega\) comprises inequalities of the form \({\sum }_{j > i}{A}_{ij}+{\sum }_{i > j}{A}_{ji}\le {v}_{i}\) where \({v}_{i}\) represents the valency of constituent atom i. These structural constraints can be modeled as a QUBO problem denoted by Qc such that \(\text{arg}{\min }_{A}{Q}_{{\rm {c}}}\in \varOmega\). This QUBO problem can be formulated as \({Q}_{{\rm {c}}}={\sum }_{i}{Q}_{i}\) where \({Q}_{i}\) is given by Eq. (2).
Similar to Bayesian optimization59, the proposed QC-based approximate optimization technique implements two phases. The primary phase involves random exploration of the chemical space to generate molecules and computing the associated objective function values. This exploration phase is performed by using a quantum annealer to solve \({\min }_{A}{Q}_{{\rm {c}}}\) to generate feasible molecules. The collected samples and the objective function values are recorded as \(D\equiv {({A}^{i},{F}_{\theta }^{i})}_{i=1,\mathrm{..}.N}\) where N indicates the number of exploration steps. The secondary phase of the proposed solution technique adopts a data-driven approach to train the surrogate model by minimizing the squared error as \({\min }_{\beta }{\sum }_{k\in D}{({F}_{\theta }^{k}-{\beta }^{{\rm {T}}}A)}^{2}\) to yield least square estimates for \(\beta\). Sampling a molecule in this phase then involves solving the QUBO \({\min }_{A}{\sum }_{i,j > i}{\beta }_{ij}{A}_{ij}+\lambda {Q}_{{\rm {c}}}\) where \(\lambda\) is a fixed multiplier that satisfies \(\lambda \gg {\Vert \beta \Vert }_{\infty }\). The obtained molecule and the observed objective value function are further appended to D. During each step of the second phase, the least square estimates are updated over dataset D, followed by solving the updated QUBO problem. This ensures that the adjacency matrix is sampled such that it approximately minimizes the surrogate model while maintaining the structural constraints. It is important to note that the QUBO problems in both phases of the proposed QC-based optimization technique can be cast as a fully connected graph. This graph can be mapped onto the physical hardware graph of the quantum annealer and is subjected to restrictions posed by the connectivity of the specific device. As the connectivity of the logical graph cast by the QUBO problems remains consistent throughout the optimization procedure, we obtain an embedding scheme for the mapping of the logical graph to the physical graph using a heuristic tool51. For a given optimization problem, the embedding scheme is determined only once and is reused over all sampling steps of the proposed QC-based approximate optimization technique.
Generating molecules
To demonstrate the viability of the proposed QC-assisted optimization technique for molecular generation, we conduct multiple computational experiments with various property targets. The conditional energy-based model trained with QC-assisted generative training is used as the structure-property relationship for inversion with the QC-based approximate optimization. For a given property target, appropriate target labels are obtained, and the corresponding conditional energy-based model is used to compute the objective function values for sampled molecules. Atomic identities of the molecules in the test set are used to search the associated chemical spaces with the proposed solution technique for molecular candidates that satisfies target property requirements. Ten exploration steps are performed for each optimization problem, followed by 90 sampling steps to minimize the associated free energy of the trained conditional energy-based model. The molecules generated with the QC-assisted optimization technique are recorded for various conditions on properties like partition coefficient and drug-likeness. Additionally, we also calculate the QED, SAS, and LogP values for all generated molecules using the RDKit package38 to validate the performance of the proposed QC-based optimization technique.
Baseline methods
To evaluate the efficiency of the structure-property relationship captured by the energy-based model, we benchmark their predictive performance against different neural network models that adopt various learning strategies and input types. The latent vector representations of the conditional energy-based models trained with both contrastive divergence (CD) learning57 and QC-based generative training are used as inputs to a single-layered neural network for predicting the corresponding property targets. CD-learning is an approximate learning approach and has been extensively used for training energy-based models60, so it is chosen for comparison against the energy-based model trained with the quantum generative approach. Additionally, three baseline models, including feedforward neural networks that use ECFP61 and MACCS62 molecular representations as input, as well as a GraphConv model46 that operates directly on molecules cast as graph-structured data to predict their properties directly, are chosen as baselines for property estimation performance.
In addition to revealing the exploration capabilities of the proposed QC-based molecular design approach in the sparsely populated chemical space, it is important to validate the targeted molecular generation. To this end, we compare the proposed method with two baseline deep learning-based approaches for molecular design, namely, conditional variational autoencoder (CVAE)63 and masked graph model (MGM)64, for benchmarking the targeted molecular generation results. CVAE is an autoencoder-based generative model that operates on the SMILES representations of molecules due to their compact line notation obtained with simple vocabulary and grammar rules65. On the other hand, MGM employs a graph neural network to operate on molecules represented as graph-structured data. CVAE imposes molecular properties onto the latent space of the autoencoder to learn a distribution of the molecular SMILES strings conditioned on their corresponding properties, while MGM captures a distribution over atoms and bonds in molecular graphs given partial ones. We also employ a graph-based genetic algorithm (GBGA)66 as a baseline owing to its ability to optimize combinatorial problems associated with targeted molecular design. In contrast to conventional genetic algorithms that manipulate SMILES notations67, GBGA performs crossover and mutation operations by altering the graph representation of molecules. Implementation details of the selected baselines for molecular design are provided in Supplementary Method 268,69,70,71.
Data availability
All data generated during this study are included in this article. The molecular data used for training along with their properties, are made available at the GitHub repository at https://github.com/PEESEgroup/qc-camd.
Code availability
The code accompanying this work is available upon request.
References
Segal, D. Materials for the 21st Century (Oxford University Press, 2017).
Virshup, A. M., Contreras-García, J., Wipf, P., Yang, W. & Beratan, D. N. Stochastic voyages into uncharted chemical space produce a representative library of all possible drug-like compounds. J. Am. Chem. Soc. 135, 7296–7303 (2013).
Achenie, L., Venkatasubramanian, V. & Gani, R. Computer aided Molecular Design: Theory and Practice (Elsevier, 2002).
Feynman, R. P. Simulating physics with computers. Int. J. Theor. Phys. 21, 467–488 (1982).
Willett, P. Genetic algorithms in molecular recognition and design. Trends Biotechnol. 13, 516–521 (1995).
Elton, D. C., Boukouvalas, Z., Fuge, M. D. & Chung, P. W. Deep learning for molecular design—a review of the state of the art. Mol. Syst. Des. Eng. 4, 828–849 (2019).
Alshehri, A. S. & You, F. Deep learning to catalyze inverse molecular design. Chem. Eng. J. 444, 136669 (2022).
Sliwoski, G., Kothiwale, S., Meiler, J. & Lowe, E. W. Computational methods in drug discovery. Pharmacol. Rev. 66, 334–395 (2014).
Pyzer-Knapp, E. O., Suh, C., Gómez-Bombarelli, R., Aguilera-Iparraguirre, J. & Aspuru-Guzik, A. What is high-throughput virtual screening? A perspective from organic materials discovery. Annu. Rev. Mater. Res. 45, 195–216 (2015).
Kim, S. et al. PubChem substance and compound databases. Nucleic Acids Res. 44, D1202–D1213 (2016).
Lehn, J. M. Dynamic combinatorial chemistry and virtual combinatorial libraries. Chem. Eur. J. 5, 307–326 (1999).
Schneider, G. Trends in virtual combinatorial library design. Curr. Med. Chem. 9, 2095–2101 (2002).
Hautier, G., Jain, A. & Ong, S. P. From the computer to the laboratory: materials discovery and design using first-principles calculations. J. Mater. Sci. 47, 7317–7340 (2012).
Hansen, K. et al. Machine learning predictions of molecular properties: accurate many-body potentials and nonlocality in chemical space. J. Phys. Chem. Lett. 6, 2326–2331 (2015).
Montavon, G. et al. Machine learning of molecular electronic properties in chemical compound space. N. J. Phys. 15, 095003 (2013).
Varnek, A. & Baskin, I. Machine learning methods for property prediction in chemoinformatics: quo vadis? J. Chem. Inf. Model. 52, 1413–1437 (2012).
Zunger, A. Inverse design in search of materials with target functionalities. Nat. Rev. Chem. 2, 0121 (2018).
Sanchez-Lengeling, B. & Aspuru-Guzik, A. Inverse molecular design using machine learning: generative models for matter engineering. Science 361, 360–365 (2018).
Venkatasubramanian, V., Chan, K. & Caruthers, J. M. Computer-aided molecular design using genetic algorithms. Comput. Chem. Eng. 18, 833–844 (1994).
Alshehri, A. S., Gani, R. & You, F. Deep Learning and Knowledge-Based Methods for Computer-Aided Molecular Design - Toward a Unified Approach: State-of-the-Art and Future Directions. Comput. Chem. Eng. 141, 107005 (2020).
Balamurugan, D., Yang, W. & Beratan, D. N. Exploring chemical space with discrete, gradient, and hybrid optimization methods. J. Chem. Phys. 129, 174105 (2008).
Gupta, A. et al. Generative recurrent networks for de novo drug design. Mol. Inf. 37, 1700111 (2018).
Segler, M. H. S., Kogej, T., Tyrchan, C. & Waller, M. P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 4, 120–131 (2018).
Gómez-Bombarelli, R. et al. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 (2018).
Blaschke, T., Olivecrona, M., Engkvist, O., Bajorath, J. & Chen, H. Application of generative autoencoder in de novo molecular design. Mol. Inf. 37, 1700123 (2018).
Sun, M. et al. Graph convolutional networks for computational drug development and discovery. Brief Bioinform. 21, 919–935 (2020).
Kang, S. & Cho, K. Conditional molecular design with deep generative models. J. Chem. Inf. Model. 59, 43–52 (2019).
You, J., Liu, B., Ying, Z., Pande, V. & Leskovec, J. Graph convolutional policy network for goal-directed molecular graph generation. Adv. Neural Inf. Process. Syst. 31, 6412–6422 (2018).
Cao, Y., Romero, J. & Aspuru-Guzik, A. Potential of quantum computing for drug discovery. IBM J. Res. Dev. 62, 1–6 (2018).
Ajagekar, A. & You, F. New frontiers of quantum computing in chemical engineering. Korean J. Chem. Eng. 39, 811–820 (2022).
Cao, Y. et al. Quantum chemistry in the age of quantum computing. Chem. Rev. 119, 10856–10915 (2019).
Montanaro, A. Quantum algorithms: an overview. npj Quantum Inf. 2, 15023 (2016).
Biamonte, J. et al. Quantum machine learning. Nature 549, 195–202 (2017).
Khatami, M. H., Mendes, U. C., Wiebe, N. & Kim, P. M. Gate-based quantum computing for protein design. PLoS Comput. Biol. 19, e1011033 (2023).
Preskill, J. Quantum computing in the NISQ era and beyond. Quantum 2, 79 (2018).
Robert, A., Barkoutsos, P. K., Woerner, S. & Tavernelli, I. Resource-efficient quantum algorithm for protein folding. npj Quantum Inf. 7, 38 (2021).
Ajagekar, A., Humble, T. & You, F. Quantum computing based hybrid solution strategies for large-scale discrete-continuous optimization problems. Comput. Chem. Eng. 132, 106630 (2020).
Landrum, G. RDKit: Open-source cheminformatics. http://www.rdkit.org (2010)
Irwin, J. J. & Shoichet, B. K. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 (2005).
Dwivedi, V. P., Joshi, C. K., Laurent, T., Bengio, Y. & Bresson, X. Benchmarking graph neural networks. J. Mach. Learn. Res. 23, 1–48 (2022).
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012).
Wildman, S. A. & Crippen, G. M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 39, 868–873 (1999).
Ertl, P. & Schuffenhauer, A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminf. 1, 8 (2009).
Cereto-Massagué, A. et al. Molecular fingerprint similarity search in virtual screening. Methods 71, 58–63 (2015).
Stepišnik, T., Škrlj, B., Wicker, J. & Kocev, D. A comprehensive comparison of molecular feature representations for use in predictive modeling. Comput. Biol. Med. 130, 104197 (2021).
Duvenaud, D. K. et al. Convolutional networks on graphs for learning molecular fingerprints. Adv. Neural Inf. Process. Syst. 28, 2224–2232 (2015).
Kadowaki, T. & Nishimori, H. Quantum annealing in the transverse Ising model. Phys. Rev. E 58, 5355–5363 (1998).
Dattani, N., Szalay, S. & Chancellor, N. Pegasus: The second connectivity graph for large-scale quantum annealing hardware. Preprint at https://arxiv.org/abs/1901.07636 (2019).
Klymko, C., Sullivan, B. D. & Humble, T. S. Adiabatic quantum programming: minor embedding with hard faults. Quantum Inf. Process. 13, 709–729 (2014).
Okada, S., Ohzeki, M., Terabe, M. & Taguchi, S. Improving solutions by embedding larger subproblems in a D-Wave quantum annealer. Sci. Rep. 9, 2098 (2019).
Cai, J., Macready, W. G. & Roy, A. A practical heuristic for finding graph minors. Preprint at https://arxiv.org/abs/1406.2741 (2014).
D-Wave. Qbsolv documentation, https://docs.ocean.dwavesys.com/projects/qbsolv (2019).
Vuffray, M., Coffrin, C., Kharkov, Y. A. & Lokhov, A. Y. Programmable quantum annealers as noisy Gibbs samplers. PRX Quantum 3, 020317 (2022).
McGeoch, C. & Farre, P. The D-wave advantage system: an overview (2020). https://www.dwavesys.com/resources/white-paper/the-d-wave-advantage-system-an-overview/.
Taylor, G. W., Hinton, G. E. & Roweis, S. Modeling human motion using binary latent variables. Adv. Neural Inf. Process. Syst. 19, 1345–1352 (2006).
Mnih, V., Larochelle, H. & Hinton, G. E. Conditional restricted boltzmann machines for structured output prediction. Proceedings of the Twenty-Seventh Conference on Uncertainty in Artificial Intelligence, 514–522 (2011).
Hinton, G. E. Training products of experts by minimizing contrastive divergence. Neural Comput. 14, 1771–1800 (2002).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2014).
Frazier, P. A tutorial on Bayesian optimization. Preprint at https://arxiv.org/abs/1807.02811 (2018).
Carreira-Perpinan, M. A. & Hinton, G. In International Workshop on Artificial Intelligence and Statistics 33–40 (PMLR).
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Durant, J. L., Leland, B. A., Henry, D. R. & Nourse, J. G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 42, 1273–1280 (2002).
Lim, J., Ryu, S., Kim, J. W. & Kim, W. Y. Molecular generative model based on conditional variational autoencoder for de novo molecular design. J. Cheminf. 10, 1–9 (2018).
Mahmood, O., Mansimov, E., Bonneau, R. & Cho, K. Masked graph modeling for molecule generation. Nat. Commun. 12, 3156 (2021).
Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988).
Jensen, J. H. A graph-based genetic algorithm and generative model/Monte Carlo tree search for the exploration of chemical space. Chem. Sci. 10, 3567–3572 (2019).
Douguet, D., Thoreau, E. & Grassy, G. A genetic algorithm for the automated generation of small organic molecules: drug design using an evolutionary algorithm. J. Comput.-Aided Mol. Des. 14, 449–466 (2000).
Dewancker, I. et al. A strategy for ranking optimization methods using multiple criteria. Workshop on Automatic Machine Learning, 11–20 (2016).
Brown, N., Fiscato, M., Segler, M. H. & Vaucher, A. C. GuacaMol: benchmarking models for de novo molecular design. J. Chem. Inf. Model. 59, 1096–1108 (2019).
Bertz, S. H. Branching in graphs and molecules. Discrete Appl. Math. 19, 65–83 (1988).
Ertl, P., Rohde, B. & Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 43, 3714–3717 (2000).
Acknowledgements
This material is based upon work supported by the National Science Foundation (NSF) under Grants Nos. 2029327 and 2229092.
Author information
Authors and Affiliations
Contributions
A.A. and F.Y. conceived the research; A.A. constructed the deep learning models and optimization methods; F.Y. validated their correctness, analyzed their performance, and supervised the work. Both A.A. and F.Y. discussed the results, wrote, and reviewed the paper’s contents and Supplementary information.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ajagekar, A., You, F. Molecular design with automated quantum computing-based deep learning and optimization. npj Comput Mater 9, 143 (2023). https://doi.org/10.1038/s41524-023-01099-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-023-01099-0
- Springer Nature Limited