Abstract
Powdery mildew is a devastating disease that affects wheat yield and quality. Wheat wild relatives represent valuable sources of disease resistance genes. Cloning and characterization of these genes will facilitate their incorporation into wheat breeding programs. Here, we report the cloning of Pm57, a wheat powdery mildew resistance gene from Aegilops searsii. It encodes a tandem kinase protein with putative kinase-pseudokinase domains followed by a von Willebrand factor A domain (WTK-vWA), being ortholog of Lr9 that mediates wheat leaf rust resistance. The resistance function of Pm57 is validated via independent mutants, gene silencing, and transgenic assays. Stable Pm57 transgenic wheat lines and introgression lines exhibit high levels of all-stage resistance to diverse isolates of the Bgt fungus, and no negative impacts on agronomic parameters are observed in our experimental set-up. Our findings highlight the emerging role of kinase fusion proteins in plant disease resistance and provide a valuable gene for wheat breeding.
Similar content being viewed by others
Introduction
Bread wheat (Triticum aestivum L.) is an important staple food crop that plays a vital role in global food security1. However, sustainable wheat production across the globe is threatened by its susceptibility to various pests and diseases. Among various diseases, powdery mildew caused by Blumeria graminis f. sp. tritici (Bgt) is a major wheat disease worldwide that affects grain yield and processing quality. This disease typically leads to yield losses higher than 1% globally and as high as 3.27% in China2. The development and deployment of resistant wheat varieties have been regarded as the most economical, effective, and sustainable way to mitigate the losses caused by powdery mildew.
To date, more than one hundred Pm resistance genes/alleles at approximately 60 loci in wheat and its wild relatives have been documented3,4; however, only a few Pm genes, including Pm2, Pm4, Pm5, Pm6, Pm8, Pm21, and Pm52, have been widely used in developing disease-resistant wheat varieties5,6,7,8. The majority of Pm genes cannot be utilized due to associated linkage drags causing adverse pleiotropism9 or narrow-spectrum resistance that often becomes ineffective with the evolution of new Bgt races10. This necessitates the identification and cloning of new genes that confer broad-spectrum powdery mildew resistance with no adverse effects on wheat agronomic characteristics.
Owing to its large genome size, cloning a gene in wheat by map-based cloning was time-consuming and challenging11. However, the availability of recently released genome sequence resources and tools, such as the genome sequences of more than ten hexaploid wheat cultivars, has largely facilitated gene mapping and cloning in wheat. Since Pm3 was isolated from wheat in 200412, 20 Pm resistance genes have been cloned. Thirteen of these 20 genes, i.e., Pm312, Pm813 (ortholog of Pm3), Pm214, Pm6015, Pm1716 (allele of Pm8), Pm2117,18, Pm4119, Pm5e20, Pm1a21, MlIW172/MlWE1822 (allelic to Pm60), Pm1223 (ortholog of Pm21), Pm6924, and Pm5525, encode nucleotide-binding leucine-rich repeat (NLR) immune receptors. Among the remaining seven Pm genes, Pm24, WTK4, and Pm36 encode tandem kinases26,27,28, Pm4 and Pm13 encode kinase fusion proteins29,30, and the two broad-spectrum resistance genes Pm38/Yr18/Lr34/Sr57 and Pm46/Yr46/Lr67/Sr55 encode an ABC transporter and a hexose transporter, respectively31,32.
Furthermore, wild relatives of hexaploid bread wheat serve as reservoirs of genetic diversity for important agronomic traits, including disease resistance genes17, and more than half of the currently designated Pm genes have been derived from secondary and tertiary gene pools of wheat33,34. Although Pm genes from wild relatives play an important role in breeding for disease resistance, it is very difficult to fine-map and clone alien genes from secondary or tertiary gene pools in the wheat background compared to those from common wheat due to homoeologous recombination suppression, lack of alien chromosome-specific markers and the unavailability of annotated reference genomes. To date, only six Pm genes have been cloned from wild relatives of wheat in secondary and tertiary gene pools including recently reported Pm5525 (Dasypyrum villosum L.) and Pm1330 (Ae. longissima); for the other four genes, Pm12 (Ae. speltoides) and Pm21 (Dasypyrum villosum L.) are orthologous, and Pm8 and Pm17 (Secale cereale L.) were cloned by Pm3 homology-based cloning. The difficulty of gene cloning hinders a better understanding of the molecular basis of these genes and limits their deployment in wheat breeding.
The powdery mildew resistance gene Pm5735 was derived from Aegilops searsii Feldman & Kislev ex Hammer (2n = 2x = 14, SsSs), an S-genome species from the section Sitopsis (Jaub. & Spach) Zhuk in the secondary gene pool of wheat. Like disease resistance gene Sr5136, Pm57 has been transferred into bread wheat from Ae. searsii. In previous study, we screened a complete set of Chinese Spring (CS)-Ae. searsii chromosome addition lines and found that the chromosome 2Ss addition line (TA3581) confers resistance to powdery mildew35. We developed ten CS-Ae. searsii 2Ss translocation lines based on homoeologous recombination induced by the pairing homeologous gene (Ph1) mutation (ph1b). Cytogenetic analyses and powdery mildew resistance assays of the translocation lines localized Pm57 to the fragment length (FL) interval of FL0.75-0.87 on the long arm of 2Ss 35. We further physically mapped Pm57 to the long arm of 2Ss in a 5.13 Mb genomic region37.
In this work, we report the map-based cloning of Pm57. Pm57 encodes a tandem kinase protein with putative kinase-pseudokinase domains followed by a von Willebrand factor A (vWA) domain, being an ortholog of Lr9 (WTK6-vWA) that mediates resistance to wheat leaf rust38. Stable Pm57 transgenic wheat lines and introgression lines exhibit high-level, all-stage powdery mildew resistance with no apparent adverse agronomic traits. Our results provide a valuable resistance gene for further elucidating the molecular mechanisms underlying wheat powdery mildew resistance and will enable the development of wheat varieties with broad-spectrum powdery mildew resistance.
Results
High-resolution mapping and map-based cloning of Pm57
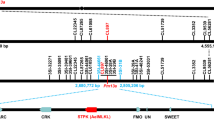
Previously, we mapped Pm57 to the long arm of 2Ss in a 5.13 Mb genomic region using an F2 population generated by crossing recombinant line 89(6)88 containing Pm57 with TA3809, a CS ph1b deletion mutant that can promote homoeologous chromosome pairing and recombination37 (Fig. 1a). To fine-map Pm57, nine heterozygous recombinant plants harboring Pm57 were self-pollinated to develop F3 populations segregated for Pm57. A total of 3,380 F3 individuals were subsequently screened using two Pm57 flanking markers (X67593 and X62492), resulting in the identification of 104 CS-Ae. searsii 2Ss recombinants (Fig. 1b). The plant responses to Bgt were subsequently assessed in the progeny of these 104 recombinants by inoculation with Bgt isolate E09 at the seedling stage in a growth chamber.
a Initial physical-mapping by comparing the chromosome recombination breakpoints of the resistant and susceptible recombinants positioned Pm57 within an interval flanked by the markers X67593 and X62492 on the long arm of chromosome 2Ss. TA3809 is a CS ph1b deletion mutant, and 89(6)88 is a CS-Ae. searsii recombinant line that contains a pair of recombined chromosomes, Ti2AS-2SsS.2SsL-2AL, that carry Pm57. b A high-resolution physical map delimited Pm57 to a 710 kb region between markers X10 and X13. Markers X67593 and X62492 were derived from RNA-seq of the Chinese Spring (CS)-Ae. searsii 2Ss disomic addition line TA3581, and markers X1-X16 were developed using the recently released genome sequence of Ae. searsii (TE01)1. Marker analysis of the 2Ss recombinants grouped them into six different types (Types I to VI), and the number of recombinants of each type is shown in brackets. The responses of each type to Bgt isolate E09 are displayed as R (resistant) or S (susceptible). c Twelve annotated genes (G1-G12) present in the mapping interval of the Ae. searsii reference genome (TE01). Two of these 12 genes (G4 and G5) encode proteins containing putative tandem kinase domains and represent wheat tandem kinase (WTK) genes. vWA: von Willebrand factor A domain.
Next, 16 2Ss-specific markers were designed within the mapping interval of Pm57 flanked by the markers X67593 and X62492 using the recently released genome sequence of Ae. searsii (TE01)1 (Supplementary Data 1). Marker analysis of the 104 CS-Ae. searsii 2Ss recombinants revealed six different types (Types I to VI). By integrating the Bgt responses of the 104 recombinants with the 16-marker analysis, we mapped Pm57 to the region between markers X10 and X13 (Fig. 1b). This region corresponds to a physical interval of 710 kb on the long arm of Ae. searsii chromosome 2Ss, which harbors 12 genes (referred to as G1-G12) based on the genome sequence of Ae. searsii1. It is worth noting that G4 and G5 encode proteins containing putative tandem kinase domains and represent wheat tandem kinase (WTK) genes (Fig. 1c)39,40. In addition, the G4 and G5 genes were absent in CS, while the other ten of the 12 genes were conserved and shared more than 80% amino acid sequence identity with the corresponding genes on Group 2 chromosomes of common wheat CS (Supplementary Table 1).
Identification of Pm57 candidate genes by MutRNA-Seq
To identify Pm57 candidates, we performed ethyl methane sulfonate (EMS) mutagenesis on the CS-Ae. searsii recombinant line 89(5)69. Line 89(5)69 contains a pair of recombined chromosomes (T2BS·2BL-2SsL) in which the long-arm terminal segment carries Pm57 of the Ae. searsii chromosome 2Ss that have been translocated to the long arms of CS 2B. Approximately 10,000 M0 seeds were treated with 0.6% EMS, and M1 seeds were harvested from approximately 1,598 surviving M0 plants. At least ten plants from each of the randomly selected 300 M1 families were screened for susceptible mutants using Bgt isolate E09. To eliminate the susceptibility caused by seed contamination or missing 2Ss, the screened susceptible mutants were verified using the 2Ss-specific molecular markers X67593 and X62492 flanking Pm57. Finally, 15 independent susceptible mutants from different M1 families were identified and further validated by Bgt isolate E09 assays in the M2 generation (Supplementary Fig. 1).
Furthermore, we performed mutant RNA-seq (MutRNA-seq) using five susceptible mutants (Mut51, Mut60, Mut141, Mut209 and Mut216) and the resistant wild-type parental line 89(5)69 (Fig. 2a). Alignment of the MutRNA-seq data to the 12 genes in the Pm57 mapping interval revealed that the G4 gene had EMS-type (G/C to A/T) mutations in all five mutants, whereas only one EMS-type mutation was found in genes G2 (in Mut209) and G8 (in Mut141), and none were found in G5, G6, G7, G9, G10, G11 or G12. The expression levels of the remaining two genes (G1 and G3) were too low to reliably call mutations in the transcriptome sequences (Fig. 2a and Supplementary Table 2). Therefore, the G4 gene, which encodes a tandem kinase-vWA domain protein, emerged as the most likely candidate for Pm57 among the 12 genes.
a Candidate gene identification by MutRNA-Seq. RNA-Seq reads from the parental line 89(5)69 and 5 susceptible EMS mutants (Mut51, Mut60, Mut141, Mut209 and Mut216) were mapped to the Ae. searsii reference genome sequence (TE01). Only G4 of the 12 candidate genes in the mapping interval carried EMS-type (G/C to A/T) mutations (black dots) in all 5 sequenced mutants. b EMS-derived loss-of-function mutants with mutations in the G4 gene sequence. Gene structure of G4 (from the start to the stop codon) was presented. Rectangles indicate coding exons, and gray straight lines indicate introns. The positions of the three conserved domains and the predicted amino acid changes caused by the EMS mutations are indicated. The two protein kinase domains and the vWA domain are shown in orange, blue, and green, respectively. “c.” for a coding DNA sequence and “p.” for a protein sequence. A frameshift mutation is detected in Mut216 with a G/A point mutation in the splice acceptor site of intron 9. The other ten mutants harbored missense or nonsense mutations.
Validation of G4 by EMS mutants, gene silencing, and transgenic assays
To validate the candidacy of G4 for Pm57, we sequenced the genomic DNA and cDNA sequences of the G4 gene and another WTK gene (G5) in the resistant 2Ss-2A recombinant line 89(5)69 as well as the 15 susceptible mutants for sequence comparison. In the resistant line 89(5)69, the G4 gene was 9473 bp in length and contained 14 exons with a coding sequence of 3489 bp (Fig. 2b); moreover, the sequence of G4 in 89(6)69 was identical to that in the reference genome TE01. Intriguingly, G4 had at least seven alternative splicing variants, designated IF1-IF7; IF1 was the main isoform, representing 50.6% (39 out of 77 tested G4 cDNA clones) at 0 h post-inoculation (hpi) with Bgt isolate E09 and increasing to 80.9% at 24 hpi; and isoforms IF2-IF7 were much less abundant (1.3–27.3%). IF1 encodes a full-length intact G4 protein with Kin I, Kin II and vWA domains, while IF2 and IF3 encode proteins with truncated Kin I and Kin II domains, and IF4-IF7 encode proteins with only a truncated Kin I domain (Supplementary Fig. 2). Gene sequence comparison revealed that 11 of the 15 susceptible mutants had SNPs in G4 that resulted in amino acid substitutions, premature stop codons, or relocation of the intron/exon splice sites (Fig. 2b). Specifically, a frameshift mutation was found in Mut216, with a G/A point mutation in the splice acceptor site of intron 9. Mut351 was the same as Mut60 in G4, both of which had a nonsense mutation that gave rise to a premature stop codon at amino acid position 1,081. The other eight mutants (G78D in Mut223, G177E in Mut141, G193R in Mut51, D209N in Mut210, G424D in Mut92, P747L in Mut121, R829W in Mut22, and G903D in Mut209) harbored missense mutations that occurred in the kinase I (Kin I), kinase II (Kin II) or vWA domains (Fig. 2b). In addition, no sequence variations in the G5 gene were found among the susceptible mutants.
Next, virus-induced gene silencing (VIGS) was performed to validate candidate genes G4 and G5. The resistant introgression line 89(5)69 inoculated with G4-VIGS constructs lost resistance to powdery mildew, whereas the plants inoculated with G5-VIGS constructs and empty vector control constructs remained resistant (Supplementary Fig. 3). These results were consistent with the conclusions drawn from the MutRNA-Seq analyses and suggested that G4 is the prime candidate for Pm57.
Transgenic complementation of the susceptible cv. Fielder was subsequently performed to confirm the role of G4 in conferring resistance against powdery mildew. We inserted the CDS (isoform IF1) of G4 into the expression vector pWMB110 driven by the maize ubiquitin (Ubi) promoter and transformed it into cv. Fielder by Agrobacterium-mediated transformation. A total of 36 T0 plants (L1 to L36) were generated, and 33 were identified as positive transgenic plants (Supplementary Fig. 4). With the exceptions of L20 and L29, all the positive T0 transgenic plants (+) were highly resistant to the Bgt isolate E09, whereas all the negative plants (−, L5, L23 and L27) were as susceptible as the WT Fielder (Supplementary Fig. 4). This difference in powdery mildew disease resistance was also observed for the T1 transgenic lines and the WT (Supplementary Table 3); 16 representative lines are shown in Fig. 3a. In addition, qRT‒PCR analyses revealed no expression of the G4 gene in the positive transgenic lines L20 and L29, explaining why these two positive lines were susceptible to powdery mildew (Fig. 3b). Taken together, the results of genetic mapping, mutant analysis, gene silencing, and transgenic assays confirmed that G4 is Pm57.
a Powdery mildew resistance assay of 16 T1 transgenic lines. Positive T1 transgenic lines (+, except L20 and L29) of G4 showed high resistance to the Bgt isolate E09. Fielder and the negative line L5 were used as susceptible controls. The L4 negative plants are the T1 individuals without G4 gene that segregated from the T0 transgenic plant L4. Three individuals of each independent line are shown. The ‘+’ and ‘−’ signs on each leaf indicate the presence or absence of the G4 gene, respectively. Scale bars = 0.5 cm. b Transcript levels of G4 in the leaves of 16 T1 transgenic lines and the Pm57 introgression line 89(5)69. The T1 line L5 was Pm57 negative, and the remaining 15 T1 lines were all positive. The first leaves of two-leaf seedlings were sampled for RNA extraction. Three biological replicates with leaves from four individual plants mixed as one replicate were used for expression analysis. Transcript levels were examined by qRT‒PCR with TaActin as an endogenous control and calculated using the comparative CT method. The data are presented as the means ± SDs from three biological replicates. WT, nontransgenic control (Fielder); N.D., not detected. *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed Student’s t test). Asterisks indicate significant differences in each transgenic line compared with the 89(5)69 control. There was no expression of the G4 gene in the positive transgenic lines L20 and L29, explaining why these two positive lines were susceptible to powdery mildew. Source data are provided as a Source Data file.
Subcellular localization and structural analysis of the Pm57 protein
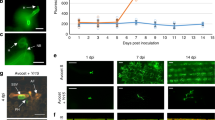
The subcellular localization of Pm57 was investigated through transient transformation of wheat leaf protoplasts. We constructed a fusion protein of Pm57 with green fluorescent protein (GFP), and the resulting Pm57-GFP construct and the GFP control were introduced into wheat protoplasts. As shown in Fig. 4a, green fluorescence of the GFP control was observed in the nucleus and cytoplasm. Similarly, the Pm57-GFP fusion protein was fluorescent in the nucleus and cytoplasm in wheat (Fig. 4a).
a Subcellular localization of Pm57 in wheat protoplasts. 35 S::GFP or 35 S::Pm57-GFP plasmids were cotransformed into wheat protoplasts with the nuclear marker plasmid AtPIF4-mCherry. The GFP, mCherry and chlorophyll fluorescence was visualized under a confocal laser-scanning microscope. 35 S::GFP served as a control. Scale bars, 10 μm. Three independent experiments were performed. b Protein structure prediction of Pm57. A three-dimensional model of Pm57 was predicted by AlphaFold. Orange, kinase domain; blue, pseudokinase domain; green, vWA domain; yellow, putative Vwaint domain. The red spheres indicate amino acid substitutions resulting from the EMS mutagenesis.
To study the structural characteristics of the Pm57 protein sequence, we compared it with the structures of eight reported WTKs (Rpg, Un8, Yr15, Sr60, Sr62, Pm24, WTK4, and Lr9)41,42. Based on the sequence conservation of the key amino acid residues in the two kinase domains43, Pm57 was classified as a tandem kinase-pseudokinase protein, similar to Yr15, Pm24, Lr9, and Sr62 (Supplementary Fig. 5). The amino acid residues in the VIb (catalytic loop) of Pm57 were LD (leucine–aspartic acid) (Supplementary Fig. 5), suggesting that Pm57 is a non-RD (non-arginine–aspartate) kinase. To explore the structure of Pm57 in detail, AlphaFold44 was used to generate a 3D model of Pm57. The tertiary structure of Pm57 is modular, with kinase-pseudokinase domains, a vWA domain, and a putative Vwaint domain, as described by ref. 38. (Fig. 4b). We also observed that the kinase domains of Pm57 were highly symmetrical, similar to those of WTK4 and Lr9 but not similar to those of the other five reported TPKs (Fig. 4b and Supplementary Fig. 6). Notably, the amino acid sequence of Pm57 was highly similar (88.3%) to that of Lr9 (Supplementary Fig. 5).
Histopathological characterization of Pm57
To evaluate the potential mechanism involved in Pm57-mediated resistance, Bgt development was evaluated in susceptible CS and resistant 89(5)69 plants. In 89(5)69, the Bgt spores germinated and developed normally from 6 to 12 hpi without histochemical differences in comparison with those of CS (Fig. 5a). In CS, Bgt gradually invaded epidermal host cells, with haustoria (Hau) visible at 24 hpi and the formation of secondary hyphae (Hyp) observed at 36 hpi. In contrast, no haustoria or hyphae were detected at 24 and 36 hpi, respectively, in 89(5)69 (Fig. 5a). Bgt-infected leaf staining by diaminobenzidine (DAB) and trypan blue (TPN) further revealed robust accumulation of H2O2 and cell death in the Bgt-interacting host cells of resistant 89(5)69 (Fig. 5). Beginning at 24 hpi, a much greater proportion of plant cells was detected to accumulate intracellular ROS (intraROS) in 89(5)69 (more than 40% of the Bgt-infected cells) than in CS (~5%) (Fig. 5a, b). Quantitative assessment of cell death at 48 hpi revealed that 37.7% of the Bgt-infected cells in 89(5)69 died, while only 3.3% of cells died in CS (Fig. 5c). These results suggest that Pm57 inhibits haustorium formation in Bgt plants and that this process might be correlated with H2O2 accumulation and induced cell death.
a, b Detection of H2O2 accumulation via DAB staining of Bgt-infected leaves. Seven-day-old plants were inoculated with the Bgt isolate E09. Staining was performed on the Bgt-infected leaves at 6, 12, 24, 36, and 48 h post inoculation (hpi). The Bgt spores germinated and developed normally from 6 to 12 hpi, and no histochemical difference was observed between 89(5)69 and CS. In CS, Bgt gradually invaded epidermal host cells, with haustoria (Hau) visible at 24 hpi and the formation of secondary hyphae (Hyp) observed at 36 hpi. In contrast, no haustoria or hyphae were detected at 24 and 36 hpi in 89(5)69, respectively. Brown staining shows the accumulation of H2O2. intraROS: intracellular ROS; scale bars, 100 μm. c Trypan blue staining of Bgt-infected leaves at 48 hpi to visualize plant cell death. Scale bars, 100 μm. Bgt-infected leaf segments were stained with Coomassie Brilliant Blue for the observation of fungal structures. The data are presented as the means ± SDs from three biological replicates. ***p < 0.001 (two-tailed Student’s t test). Source data are provided as a Source Data file.
qRT‒PCR analyses demonstrated that Pm57 was more highly expressed in wheat leaf tissues than in roots and stems (Supplementary Fig. 7a). Upon pathogen infection with the Bgt isolate E09, the expression of Pm57 increased 4-fold and peaked at 12 hpi in the leaves of 89(5)69 seedlings, after which the expression slightly decreased at 48 hpi (Supplementary Fig. 7b). In addition, the upregulation of pathogenesis-related (PR) genes (i.e., PR1, PR2, PR3, PR4, and PR9) upon inoculation with the Bgt isolate E09 was significantly greater in the leaves of the transgenic lines than in those of the susceptible control Fielder, and the expression levels of the PR genes peaked at timepoints later than those of Pm57 (Supplementary Figs. 7b and 8). These expression patterns of the Pm57 and PR genes imply that powdery mildew resistance of Pm57 may involve PR genes up-regulated expression.
Evolutionary analysis of Pm57
A comparative analysis of published wheat genome sequences showed synteny is conserved largely in tribe Triticeae for Pm57 mapping genomic regions (Supplementary Fig. 9); however, Pm57 orthologs were present only in Aegilops bicornis (TB01) (SbSb), Ae. longissima (TL05) (SlSl), Ae. speltoides (TS01) (SS), and the B subgenome of T. dicoccoides (WEWSeq v1) (AABB), and the proteins encoded by these orthologous genes had 81.8–87.8% sequence similarity with Pm57 (Supplementary Figs. 9 and 10). Notably, Pm57 is absent in the wheat reference genome sequence of cv. Chinese Spring, but sequences highly similar to that of Pm57 were present in the syntenic region of several bread wheat cultivars (Supplementary Data 2). These highly similar sequences were manually annotated and shared more than 80% amino acid sequence identity with Pm57. Synteny analysis was also performed for the flanking genomic regions around the Pm57 locus in Ae. searsii (TE01) and the Lr9 donor Ae. umbellulata (TA1851). The adjacent genomic region of Pm57 was syntenic to the Lr9 genomic region in Ae. umbellulata (Supplementary Fig. 11 and Supplementary Data 3), suggesting that Pm57 may be an ortholog of Lr9. Lr9-vs.-Pm57 reciprocal BLAST was performed between the Ae. searsii (TE01) and Ae. umbellulata (TA1851) assemblies, and the best BLAST hit with the Lr9 genomic sequence was Pm57, while the best BLAST hit with the Pm57 genomic sequence was Lr9 (Supplementary Table 4). Therefore, genome similarity, gene collinearity and “reciprocal best hits” analyses revealed the Pm57 gene to be an ortholog of Lr9; thus, this gene was designated WTK6b-vWA.
To gain insight into the evolution of Pm57 and Lr9, we calculated the ratio (Ka/Ks) of the nonsynonymous substitution rate (Ka) to the synonymous substitution rate (Ks) for both Pm57 and Lr9. The Ka/Ks ratio revealed that although WTK6 (Pm57 vs. Lr9) primarily underwent strong purifying selection (Ka/Ks = 0.42 < 1), the three domains presented diverse selective pressures (Supplementary Table 5). Strikingly, pseudokinase (K2) exhibited relaxed constraints or neutral evolution (Ka/Ks = 0.77), whereas the kinase (K1) and vWA domains exhibited strong purifying selection (Ka/Ks = 0.14 and 0.19).
To evaluate the evolutionary origin of the protein kinase domains in Pm57, each of the two kinase domains in Pm57 was queried against the Hidden Markov models (HMMs) of 157 protein kinase subfamilies developed by Legti-Shiu and Shui45. Both protein kinase domains in Pm57 were assigned to the protein kinase subfamily DLSV (DUF26, SD-1, LRR-VIII and VWA) in the receptor-like kinase (RLK)/Pelle family. Therefore, Pm57 has a DLSV-DLSV configuration similar to that of Rpg1, Pm24, and Sr6246. Notably, a portion of the DLSV members were subsequently recognized as the LRR_8B subfamily (cysteine-rich receptor-like kinases)47. To further classify the two protein kinase domains of Pm57, we used the 182 protein kinase domains identified by Klymiuk et al.43. to construct a neighbor-joining (NJ) phylogenetic tree. As shown in Supplementary Fig. 12, both of the protein kinase domains in Pm57 were assigned to the LRR_8B subfamily.
To test whether the Pm57 resistance allele is present in other wheat germplasms, an STS-Pm57 marker with a 57 °C annealing temperature was used to amplify a 530 bp genomic sequence of the Pm57 gene from 71 wheat accessions (including T. urartu, T. boeoticum, Ae. tauschii, T. monococcum, T. dicoccoides, T. durum, T. aestivum ssp. yunnanese, T. aestivum ssp. macha, T. aestivum ssp. spelta, T. aestivum ssp. tibetanum accessions and Chinese common wheat landraces and modern cultivars; Supplementary Data 4). The amplified fragment of Ae. searsii (TE01) was identical to that of CS-Ae. searsii chromosome 2Ss introgression line 89(5)69, but no identical fragment was obtained for the remaining 69 accessions (Supplementary Fig. 13). These results indicate that the STS-Pm57 marker can be used as a diagnostic marker for the effective detection of Pm57 in wheat breeding programs.
Evaluation of Pm57 application in wheat breeding
Our previous studies showed that Pm57 confers high resistance to mixed Bgt isolates collected in Henan Province37. To further study the spectrum of resistance offered by Pm57 against genetically divergent Bgt isolates, we collected 29 Bgt isolates from major wheat-growing regions of China and used them for Bgt resistance assays of both the CS-Ae. searsii introgression line 89(5)69 and the transgenic line L1. As a result, plants of both 89(5)69 and the transgenic line L1 exhibited high resistance to all 29 divergent Bgt isolates, with an infection type ranging from 0–1 (Supplementary Table 6). Furthermore, we found that 89(5)69 and positive transgenic plants exhibited high resistance from the seedling stage to the adult stage (Supplementary Fig. 14). In addition, all of the F1 plants derived from the CS-Ae. searsii recombinant (88R-3-19-1), which has a small 2Ss segment harboring Pm57, crossed with 22 wheat varieties were highly resistant to powdery mildew, indicating that Pm57 could play a role in resistance in diverse genetic backgrounds (Supplementary Fig. 15).
To determine the value of Pm57 in wheat breeding, we evaluated the major agronomic traits of the Pm57 transgenic wheat lines and introgression lines. No significant differences were observed for heading date, plant height, tiller number, thousand-grain weight or spike morphology between the three transgenic lines and the control lines, which included both the negative lines and the WT (Fig. 6 and Table 1). Additionally, there was no significant difference between the introgression lines (Type IV-1 and Type IV-2; in Fig. 1b, 5.57–9.12 Mb) and the parental line TA3809 for these traits (Fig. 6 and Table 1). The results showed that Pm57 had no obvious adverse effects on important agronomic traits.
a Whole-plant, spike, and seed growth habits of Fielder (−) and Pm57 transgenic plants (+). b The spikes and seeds of TA3809 and the CS-Ae. searsii Pm57 introgression lines. Types IV-1 and IV-2 are two CS-Ae. searsii introgression lines with small 2Ss segments carrying Pm57 (Type IV in Fig. 1b; 2Ss fragment sizes between 5.57 Mb and 9.12 Mb, ~1% of the 2Ss chromosome). TA3809 served as a control for these introgression lines.
Discussion
The development of wheat-alien recombinants is not only a tool for cloning alien genes but also one of the best approaches for transferring favorable genes from wild relatives to increase the genetic diversity of cultivated wheat. However, homoeologous recombination between wheat and alien chromosomes is suppressed by the pairing of homoeologous (Ph) genes, hampering the development of recombinants with smaller alien segments, which is essential for mapping and deploying exotic wheat genes in bread wheat cultivars48. In this study, we used CS-Ae. searsii 2Ss F3 recombinants developed from a cross between the Ph1 locus deletion mutant TA3809 and the powdery mildew-resistant translocation line 89(6)88 to fine-map the broad-spectrum powdery mildew resistance gene Pm57. The Pm57 candidate region harbored 12 genes in the Ae. searsii reference genome assembly (TE01)1. We further performed MutRNA-Seq to narrow the Pm57 candidate genes in the target mapping interval and identified G4 as the most likely candidate for further transgenic validation. Unexpectedly, the CDS of G4 was incompletely annotated as EVM0016946 in the Ae. searsii reference genome; this gene encodes a protein with a single kinase domain followed by a vWA domain. We obtained the full-length sequence of G4 from Pm57 direct donor RNA-seq data37 and aligned the MutRNA-Seq data from the susceptible mutants against the G4-corresponding unigenes. In recent years, MutRNA-Seq has been proven to be an effective method for cloning disease resistance genes; for example, Sr6246 and Lr9/Lr5838 were cloned by exploiting similar MutRNA-Seq approaches.
Recently, tandem kinase proteins (TKPs) have emerged as prominent new components of disease resistance in Triticeae39. To date, nine TKPs, namely, Rpg149, Yr15 (WTK1)43, Sr60 (WTK2)50, Pm24 (WTK3)27, WTK426, Sr62 (WTK5)46, Lr9 (WTK6-vWA)38, Rwt451, and Pm36 (WTK7-TM)28, have been identified to confer resistance against various fungal pathogens. Pm57 (WTK6b-vWA) from Ae. searsii is an additional member of the TKP family that confers resistance to wheat powdery mildew. It contains tandem kinase domains and a Willebrand factor A (vWA) domain and is the second identified protein with this type of unusual WTK-vWA structure following Lr9, which confers resistance to wheat leaf rust. The vWA/Vwaint domains are present in bacteria, archaea and eukaryotes and are considered to participate in protein‒protein interactions52,53. For example, the vWA domain of the human copine protein can interact with a wide variety of signaling molecules containing coiled-coil domains54. In plants, vWA-containing copine proteins were shown to be regulators of basal and R-mediated disease resistance, suggesting that vWA-containing proteins may play an important role in plant disease resistance55. In this study, eight susceptible mutants harbored missense mutations in the Kin I, Kin II or vWA domains (Fig. 2), indicating that each domain of Pm57 is essential for resistance to the Bgt pathogen. A similar phenomenon was also observed for the Rpg1, Pm24, Sr62 and Lr9 genes27,38,46,49. Neither sequence variation nor changes in the expression level of Pm57 were found in four of the 15 susceptible mutants (Supplementary Fig. 16), providing a resource to further study other genes or elements required for Pm57 function.
Comparative analysis of Pm57 orthologs among plant kingdoms revealed that its orthologs are present only in Ae. bicornis (TB01.2S01G0903000.1), Ae. longissima (TL05.2S01G0920200.1), Ae. speltoides (TS01.2B01G0896900.1), T. dicoccoides (TRIDC2BG085800.1) and several sequenced bread wheat accessions (Supplementary Fig. 9 and Supplementary Data 2), suggesting a likely recent origin of Pm57 after the divergence of Triticeae species and the reticulate evolutionary nature of wheat56,57. However, several genes encoding proteins with a single kinase domain followed by a vWA domain were identified not only in Triticeae species but also in other species (Supplementary Data 2). Moreover, the sequences of single kinase-vWA proteins from Ae.bicornis.TB01.Un01G0378900.1 and Thint.05G0470400.1.p are very similar with Pm57 (>90%, Supplementary Fig. 17). These results suggest that the tandem kinase-vWA protein Pm57 is likely derived from a single kinase-vWA protein. Previous studies have shown that tandem kinase proteins may be derived from the duplication or fusion of two kinase domains. If two kinase domains of the same gene show high sequence similarity and originate from the same kinase family or subfamily, they are derived from gene duplication; if two kinase domains originate from two different kinase families or subfamilies or share relatively low similarity, they are derived from gene fusion43. The two kinase domains of Pm57 shared a relatively high similarity (58.04%) in amino acid sequence, and both were classified into the LRR_8B subfamily47 and within the same group with two kinase domains of Lr9 (Supplementary Fig. 12). These results indicated that the two kinase domains of Pm57 could have resulted from a duplication event.
Although Pm57 was inferred to be an ortholog of Lr9, with a high protein sequence similarity of 88.3%, the amino acid sequences in the pseudokinase and vWA domains and protein structures are clearly different between Pm57 and Lr9 (Supplementary Fig. 5 and 17). The overlap analysis of the predicted protein structures revealed that the overlap of Pm57 and Lr9 was very high in kinase domains but poor in the disordered regions of the pseudokinase domain and the vWA domain (Supplementary Fig. 18). Further comparison of the overall disorder of Pm57 and Lr9 revealed that the disorder of Pm57 was greater than that of Lr9, especially in the pseudokinase domain and vWA domain regions (Supplementary Fig. 19). In addition, although purifying selection was observed for both Pm57 and Lr9, the selection pressures applied to the kinase and pseudokinase domains markedly differed (Supplementary Table 5). Wang et al. hypothesized that the pseudokinase and vWA domains of Lr9 might serve as integrated decoys for the detection of pathogen effectors38. It is reasonable to speculate that the differences in pseudokinase and vWA domains between Pm57 and Lr9 may lead to differences in the detection of Bgt and Pt pathogen effectors and thus confer resistance to powdery mildew and leaf rust, respectively. A similar case was recently described for WTK3 and RWT4, which showed high sequence similarity but conferred resistance to powdery mildew and wheat blast, respectively27,51. WTK3 and RWT4 were considered alternative alleles of the same gene with different functional specificities51. This phenomenon of functional specificity of orthologous genes or alleles is very worthy of further study to better elucidate the resistance mechanism of resistance genes to pathogens.
In summary, we cloned Ae. searsii-derived Pm57, which confers broad-spectrum and all-stage resistance against Bgt. Pm57 encodes a protein consisting of tandem kinase domains and a vWA domain (WTK6b-vWA). Since Pm57 is an ortholog of Lr9, our results provide evidence for the resistance of orthologous genes or alleles to different diseases. The isolation of Pm57 lays a solid foundation for further understanding the molecular mechanism underlying WTK-vWA-mediated resistance to various plant diseases. The identified introgression lines harboring Pm57 in a small alien segment, as well as diagnostic function markers, will facilitate the improvement of elite wheat varieties with durable powdery mildew resistance.
Methods
Plant materials
The common wheat (T. aestivum L.) cultivar Chinese Spring (CS, TA3808), the CS ph1b mutant stock (TA3809), and the CS-Ae. searsii chromosome 2Ss recombinant lines 89(5)69 (TA5109) and 89(6)88, both of which carry Pm57, were used for Pm57 mapping and cloning in this study (Supplementary Table 7). The introgression line 89(6)88 was crossed with TA3809 to produce F237 and F3 populations with homozygous ph1b and segregated for the recombinant chromosome 2AS-2SsS ∙ 2SsL-2AL carrying Pm57. The CS-Ae. searsii 2Ss recombinants identified in this study were generated from F2 populations using the Pm57 flanking markers X67593 and X62492. Line 89(5)69 was used for Pm57 gene cloning and expression analyses. The wheat cultivar Fielder was used for wheat protoplast preparation and genetic transformation. In addition, two Pm57 introgression lines, Type IV-1 and Type IV-2, developed in this study were used to investigate the effect of Pm57 on the main agronomic traits. The CS-Ae. searsii Pm57 recombinant 88R-3-19-137 was crossed with 22 wheat varieties to determine whether Pm57 can confer powdery mildew resistance in diverse genetic backgrounds (Supplementary Table 7). A total of 71 wheat accessions, including diploid, tetraploid and hexaploid wheat accessions, were employed to investigate the presence of the Pm57 resistance allele in other wheat germplasms (Supplementary Data 4). TA3808 and TA3809 were obtained from the Wheat Genetics Resource Center (WGRC) at Kansas State University, USA, and the CS-Ae. searsii 2Ss recombinants TA5109 and 89(6)88 were developed by our laboratory and provided to WGRC. The 71 wheat accessions, except 89(5)69, were kindly provided by Prof. Zhongfu Ni at China Agricultural University, China. All materials were maintained at the experimental station of Henan Agricultural University, China.
Phenotypic response to powdery mildew
The powdery mildew (Bgt) isolate E09 and 28 other genetically divergent isolates (Supplementary Table 6) collected from different regions of China were used to evaluate the powdery mildew resistance spectrum of Pm57, while the isolate E09 was used for all other resistance assays in this study. All the seedlings to be inoculated were grown in 7 × 7 cm nutrient pots under aseptic conditions. When the first leaves were fully unfolded, the seedlings were inoculated with Bgt isolates and grown in a greenhouse maintained at 18–24 °C, 16 h light/8 h dark cycle and approximately 70% relative humidity37. Disease symptoms were recorded 7 days post-inoculation (dpi) using a scale from infection type 0 to 4 (IT 0 for no visible symptoms, IT 0; for hypersensitive necrotic flecks, IT 1-4 for highly resistant, moderately resistant, moderately susceptible and highly susceptible)58. Based on the IT scores, the tested plants were classified into two groups: resistant (R, IT 0-2) and susceptible (S, IT 3-4). CS was used as a susceptible control and for propagating Bgt isolates.
The resistance of the CS-Ae. searsii chromosome 2Ss recombinants, EMS-induced susceptible mutants and Pm57 T0 transgenic plants was determined by inoculation with E09 and further confirmed using their self-pollinated progenies. If 10-15 progenies from each line were all resistant or segregated for resistance, the parental plant was considered resistant; however, if all the progenies were susceptible, the parental plant was considered susceptible. To evaluate the Pm57 resistance spectrum, four seedlings of CS-Ae. searsii chromosome 2Ss introgression line 89(5)69 and Pm57 transgenic plants were inoculated with each isolate from a total of 29 Bgt isolates in triplicate assays. To investigate the potential influence of diverse genetic backgrounds on Pm57 resistance, F1 hybrids derived from crosses of the Pm57 introgression line 88R-3-19-1 with 22 elite wheat varieties were subjected to Bgt E09 response assays, and four seedlings from each of the hybrids were subjected to each of three independent assays.
The powdery mildew resistance of primary transgenic plants (T0) was tested using detached leaves. Briefly, T0 seedlings were transplanted into plastic pots containing mixed soil composed of sand, peat, and perlite (1:1:1, v/v/v) and subsequently grown in a growth chamber for 14 days under normal growth conditions. Then, three detached leaves from each T0 plant, susceptible control Fielder and resistant control recombinant 89(5)69 were placed on Phytagar media (0.5% Phytagar; 30 ppm benzimidazole), inoculated with Bgt isolate E09 and cultured in a greenhouse. ITs were recorded 7 days after inoculation. Additionally, assessments of powdery mildew reactions in T1 transgenic plants during the whole growth period were conducted through natural transmission of the Bgt pathogen in the greenhouse. Approximately 12 plants of each T1 transgenic line were potted. Simultaneously, the highly susceptible CS was inoculated to spread the Bgt pathogen in the greenhouse.
Physical mapping of Pm57
The wheat-Ae. searsii 2Ss recombinant population was developed from the cross of the CS-ph1b mutant stock TA3809 and 89(6)8837. Nine heterozygous resistant CS-Ae. searsii 2Ss recombinants (88R-2-32-3, 88R-4-30-2, 88R-5-28-4, 88R-2-9-2, 88R-4-5-4, 88R-4-18-3, 88R-5-1-4, 88R-2-22-3, and 88R-3-6-2) were self-pollinated to produce a secondary mapping population for further mapping of Pm57. CS-Ae. searsii 2Ss recombinants in the 5.13 Mb segment harboring Pm57 were identified with the Pm57 flanking markers X67593 and X62492. The Ae. searsii (TE01)1 and Chinese Spring (v2.1)59 genome sequences were used for the development of STS-PCR markers. All the markers (Supplementary Data 1) were designed using DNAMAN 7 software (Lynnon Biosoft, San Ramon, CA, USA). DNA was extracted using the CTAB method, and genotyping of the recombinants was performed in 15 µL volumes containing 200 ng template gDNA. PCR was performed using 2x ES Taq MasterMix (CWBIO, China) under the following conditions: 94 °C for 3 min; 34 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s; and 72 °C for 3 min.
Mutant development and MutRNA-Seq
Seeds of 89(5)69 were soaked in distilled water for 6 h and treated for 16 h with a 0.6% (v/v) EMS solution with shaking at 150 rpm at room temperature. The solution was then removed, and the treated seeds were rinsed with running water for 6 h. The mutagenized M0 seeds were planted in the field, and the M1 seeds from each M0 plant were harvested at maturity. The M1 seedlings (10-15 plants for each family) were phenotyped with Bgt isolate E09 in a greenhouse. Susceptible M1 plants were advanced to the M2 generation, and 10-15 M2 seedlings were tested to confirm their susceptibility.
MutRNA-Seq was performed as described by ref. 46. Specifically, five susceptible M2 mutants (Mut51, Mut60, Mut141, Mut209 and Mut216) derived from independent M1 families as well as the resistant wild-type parental line 89(5)69 were selected for RNA-seq. The first leaves from five plants of each genotype were sampled for RNA extraction at 5 dpi with Bgt. Total RNA was extracted using TRIzol reagent (TransGen, Beijing, China). RNA-seq was performed by Annoroad Gene Technology Co., Ltd. (Beijing, China). The Illumina HiSeq X Ten platform (Illumina, USA) was used, and 39.0, 58.8, 37.8, 44.6, 52.4 and 46.1 million 150 bp paired-end clean reads were produced for 89(5)69, Mut51, Mut60, Mut141, Mut209 and Mut216, respectively. The clean reads were subsequently mapped to the Ae. searsii genome assembly (https://ngdc.cncb.ac.cn/gwh/Assembly/24532/show) using HISAT2 (version 2.0.5, default parameters). The alignment results were further processed into BAM format using SAMtools (version 1.9) with default parameters. Gene expression levels were quantified using the featureCounts tool (version 1.4.4; parameters: -T 10 -t exon -g gene_id). Based on the alignment results, we conducted single nucleotide polymorphism (SNP) calling using BCFtools (version 1.7). Briefly, SNPs were detected using the commands bcftools mpileup (with the parameters “-A -C50 -Q20 -q30 -Ou -r -f”) and bcftools call (with the parameters “-vmO z -V indels -o”). The SNPs were filtered according to the following set of read characteristics: SNP quality (QUAL < 30), genotype (GT = 0/1), low read depth (DP < 10), mapping quality (MQ < 40), non-EMS-induced point mutations (A/T to G/C transitions), and nucleotide positions with more than 2 samples supporting alternative alleles. These SNPs were excluded from further analysis.
Gene cloning and sequence analysis
TRIzol reagent (TransGen, Beijing, China) was used for RNA extraction, and 2 μg of total RNA was used for cDNA synthesis using a HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, Nanjing, China). The full-length genomic DNA (gDNA) and cDNA sequences of G4 from 89(5)69 and each of the 15 susceptible mutants were amplified using the primers listed in Supplementary Data 1. PCRs were performed in 30 µL volumes using high-fidelity Primestar polymerase (TaKaRa, Dalian, China). The PCR conditions were previously described60. Specifically, 98 °C for 3 min; 34 cycles of 98 °C for 15 s, 60 °C for 20 s, and 72 °C for 3 min; and 72 °C for 3 min. The PCR products were sequenced by the Sanger dideoxy DNA sequencing method. The sequences of G4 in each mutant and in the wild-type 89(5)69 were compared using DNAMAN 7 software.
Alternative splicing variant analysis
RNA was isolated from 89(5)69 leaf tissue inoculated with Bgt isolate E09 at 0 and 24 hpi. The primers 1 F/3R1 for the full-length G4 were used to clone alternative splicing variants from cDNA. The PCR products were purified and cloned and inserted into the pEASY-Blunt cloning vector (TransGen, Beijing, China). A total of 77 and 89 colonies at 0 and 24 h post inoculation (hpi), respectively, were selected for Sanger sequencing.
Virus-induced gene silencing
Virus-induced gene silencing (VIGS) was performed as previously described57. In brief, the plants were grown in a growth chamber under 16-h light (24 °C)/8-h dark (18 °C) and approximately 70% relative humidity. To develop specific VIGS targets, we blasted the sequences of the G4 and G5 genes against the CS and Ae. searsii genome sequences. The 3’ ends of the G4 and G5 fragments, 200-250 bp in length and with low similarity (identity <50% and stretches of 100% nucleotide identity<21 nt) with other genes, were selected as targets; these fragments were separately cloned and inserted into the BSMV-γ (BSMV, barley stripe mosaic virus) vector, resulting in the construction of γ-G4 and γ-G5. Equimolar amounts of in vitro transcripts of BSMV-α, BSMV-β and γ-G4 or γ-G5 were mixed to inoculate the fully expanded second leaves of 89(5)69 seedlings, and leaves infected with BSMV-TaPDS and BSMV-γ (empty vector) were used as controls61. Approximately 14 days after viral infection, the 3rd and 4th leaves with symptoms of viral infection were sampled for Bgt inoculation and gene silencing expression analyses, respectively. For Bgt inoculation, the samples were placed on 1% agar plates supplemented with 20 mg/mL 6-phenyladenine (6-BA) and subsequently inoculated with Bgt. After seven days, the powdery mildew resistance phenotype was evaluated. The experiments were repeated three times.
Gene expression analysis
For the gene expression analyses of G4 and G5 in the VIGS experiments, the 3rd and 4th leaves were mixed together as one sample, and three technical replicates were performed for each sample. RNA extraction and cDNA synthesis were performed as described above. Quantitative RT‒PCR (qRT‒PCR) analysis was carried out in a reaction volume of 20 µL using SYBR Mix (TaKaRa, Dalian, China) on a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). The conditions for qRT‒PCR were as follows: denaturation at 95 °C for 4 min, followed by 40 cycles of 94 °C for 20 s, 60 °C for 20 s and 72 °C for 20 s. Wheat TaActin was used as an internal reference gene27. The transcript levels were calculated using the comparative CT method62. The primer pairs 3 F/2 R and M-5F/5 R were used to determine the expression of G4 and G5, respectively (Supplementary Data 1).
To study the tissue-specific expression of Pm57, seedling roots and leaves from the three-leaf period and adult plant roots, stems, and flag leaves from the heading stage were obtained from 89(5)69. Three biological replicates with tissues from four individual plants mixed as one replicate were used for expression analysis. For expression analysis of Pm57 and the pathogenesis-related (PR) genes PR1, PR2, PR3, PR4 and PR9 in 89(5)69, the transgenic lines (L1 and L16), and Fielder, the first leaves at the two-leaf stage were used to extract total RNA before inoculation (0 h) and at 12, 24, 36, and 48 hpi with the Bgt isolate E09; for each time point, qRT‒PCR was performed on three biological replicates, with the first leaves from four individual plants mixed as one replicate. The RNA extraction, cDNA synthesis and qRT‒PCR conditions were the same as those above. The primers used to evaluate the transcript levels of the Pm57 and PR genes are listed in Supplementary Data 1. The wheat TaActin gene was used as the endogenous control. The comparative CT method was used to quantify relative gene expression.
Wheat transformation
The full-length coding sequence of G4 from 89(5)69 was inserted into pWMB110 vectors using the restriction enzymes BamH I under the control of the maize ubiquitin (Ubi) promoter. Wheat transformation was performed using the Agrobacterium-mediated method with strain EHA105 and calluses induced from cv. Fielder immature embryos63. To determine positive transgenic events, DNA was extracted from leaves of independent T0 plants, and specific PCR primers (3 F/2 R) were designed to amplify a 344 bp fragment of the G4 gene. qRT‒PCR analysis was performed to evaluate the expression levels of G4 in the first leaves of transgenic wheat plants in the T1 generation. The first leaves of four seedlings at the two-leaf stage were sampled for each line to extract RNA. RNA extraction, cDNA synthesis, and qRT‒PCR analysis were performed as above. All the reactions were performed in triplicate. The disease responses of the transgenic plants to powdery mildew were tested as described above.
Subcellular localization analysis
To determine the subcellular location of Pm57, the coding sequence of Pm57 was cloned into pJIT163-GFP vector, in which the expression of Pm57-GFP was driven by the CaMV35S promoter. An empty vector, pJIT163-GFP, was used as the negative control. The constructs were delivered into wheat protoplasts via PEG-mediated transformation according to a prior study64. Under an induction of 40% PEG-4000, control or recombinant plasmids (10 μg per construct) were co-transformed into wheat protoplasts with nucleus marker plasmid AtPIF4-mCherry65. The transformed protoplasts were cultured at 25 °C for 16 h under dark conditions, and observed using a laser confocal microscope (A1F, Nikon, Tokyo, Japan). Image acquisition was conducted with the NIS-Elements Viewer Imaging Software (version 5.21.00). The subcellular localization of Pm57 protein was determined at three times.
Pm57 protein 3D modeling prediction
To predict the 3D structure of Pm57 and other WTKs, we used the open-source code of AlphaFold v2.144. We input the amino acid sequence of each WTKs into AlphaFold v2.1, and obtained five unrelaxed, five relaxed and five ranked models in.pdb format. Among the output models, the ranked_1.pdb model had the highest confidence with the best Local Distance Difference Test (lDDT) score were utilized. The structural graphics and the positions of amino acid substitutions were visualized using PyMOL (v. 2.6.0a0).
Detection of H2O2 accumulation and plant cell death
To detect the accumulation of H2O2, the first leaves cut from 89(5)69 and CS at 6, 12, 24, 36, and 48 h post inoculation (hpi) were immediately incubated in a 3, 3’-diaminobenzidine (DAB) solution (1 mg/mL, pH 5.8) for 12 h at 25 °C, and then bleached in absolute ethanol. Before assessing the accumulation of H2O2, the bleached leaves were incubated in a 0.6% (w/v) Coomassie blue solution for 10 s and then washed with water. To detect plant cell death, the primary leaves from the 89(5)69 and CS at 48 hpi were incubated in a 0.4% Trypan blue solution for 1 min in boiling water, washed with sterile water, bleached for 16 h in chloral hydrate solution (2.5 g/mL), fixed for 20 min in ethanol-acetic acid 3:1 (v/v), and stained in a 0.6% (w/v) Coomassie blue solution for 30 s. The treated leaves were viewed under a microscope (Olympus BX53). The ratios of ROS and cell death were calculated as described by Li et al.66. Specifically, three independent experiments were performed, and three leaves were collected as biological replicates for each time, and at least 100 infected cells per leaf were observed to identify the ROS and cell death responses.
Collinearity analysis, homology searching and phylogenic analysis
Collinearity analysis among different species or subgenomes was performed using the online tool Triticeae-GeneTribe with default parameters67. The WheatOmics 1.0 (http://wheatomics.sdau.edu.cn/) and Phytozome v13 database (https://phytozome-next.jgi.doe.gov/) were used to find proteins similar to Pm57 in plant genomes68,69. Pm57 were used as queries for BLAST analysis with default parameters. All the retrieved proteins were scanned by website pfam 35.0 (http://pfam.xfam.org/) in batch mode with an E value of 0.01, and the retrieved proteins with kinase domain and vWA domain were selected. For kinase domain analysis, the 182 putative kinase or pseudokinase domains used for phylogenetic analysis of WTK1 (Yr15)43 and WTK3 (Pm24)27 were also used in this study. In addition, the proteins with a tandem kinase -vWA structure were included in the phylogenetic analysis of kinase domains. Multiple sequence alignments were carried out with ClustalW software with default settings70. The conserved motifs in the Kin I and Kin II domains were previously annotated by Klymiuk et al.43. Classification of Pm57 to protein kinase subfamily was performed by HMMER v3.3.2 (http://hmmer.org/) with HMM models developed by Legti-Shiu and Shui45. The classification was further confirmed by a neighbor-joining (NJ) phylogenetic analysis, which was conducted using MEGA7.071 with Poisson model and 1000 repetitions of bootstraps. Lr9-vs-Pm57 reciprocal BLAST is performed between Ae. searsii (TE01) and Ae. umbellulata (TA1851) assemblies by BLASTN 2.13.0+ software with default settings. The Ka (non-synonymous substitution) and Ks (synonymous substitution) substitution rates was calculated to estimate the selection pressure between Pm57 and Lr9 genes using KaKs_Calculator 3.072.
Agronomic trait evaluation of Pm57 transgenic lines and introgression lines
Three T2 transgenic lines and two introgression lines of Pm57, and the control lines were planted in the experimental fields of Henan Agricultural University (Zhengzhou, China) using a randomized block design with three replications. Each plot consisted of two 1.5-m rows spaced 25 cm apart, and 20 seeds were sown in each row. Field managements including irrigation, fertilization, herbicide and pesticide applications strictly followed local practice.
Agronomic traits were phenotyped as previously described73. In brief, for each replicate, five plants for each plant material are selected to measure agronomic traits that includes heading date (the date from sowing to the date when half of the spikes had emerged from the flag leaf of the main tiller), plant height (the length from the ground to the tip of the main spike excluding awns), spike length (the length from the rachi base to the uppermost spikelet top excluding awns), spikelet number per spike, seeds per spike and one-thousand-grain weight. The significance of differences among means of agronomic traits was determined using two-tailed Student’s t-test.
Prediction of the disorder of Pm57 and Lr9
The predisposition of Pm57 and Lr9 to a disordered structure was assessed by the Predictor of Naturally Disordered Regions (PONDR; http://www.pondr.com/) server with default settings.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data supporting the findings of this work are available within the paper and its Supplementary Information files. Detailed sequence data of Pm57 gene was deposited in NCBI database under accession OQ675542. The MutRNA-Seq data derived from the WT and five mutant plants have been deposited in NCBI’s Sequence Read Archive (SRA) under accession number PRJNA947672. Public databases WheatOmics 1.0 (http://wheatomics.sdau.edu.cn/) and Phytozome v13 (https://phytozome-next.jgi.doe.gov/) were used in this study. Source data are provided with this paper.
References
Li, L. et al. Genome sequences of five Sitopsis species of Aegilops and the origin of polyploid wheat B subgenome. Mol. Plant 15, 488–503 (2022).
Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019).
Hafeez, A. N. et al. Creation and judicious application of a wheat resistance gene atlas. Mol. Plant 14, 1053–1070 (2021).
McIntosh, R.A., Dubcovsky, J., Rogers, W.J., Xia, X.C. & Raupp, W.J. Catalogue of gene symbols for wheat - 2020 Supplement. In: GrainGenes Database (https://wheat.pw.usda.gov/GG3/wgc) (2020).
Gao, H. et al. Identification of the powdery mildew resistance in Chinese wheat cultivar Heng 4568 and its evaluation in marker-assisted selection. Front. Genet. 13, 819844 (2022).
Xu, H. et al. Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor. Appl. Genet. 128, 2077–2084 (2015).
Zhao, Z. et al. Genetic analysis and detection of the gene MlLX99 on chromosome 2BL conferring resistance to powdery mildew in the wheat cultivar Liangxing 99. Theor. Appl. Genet. 126, 3081–3089 (2013).
Nematollahi, G., Mohler, V., Wenzel, G., Zeller, F. J. & Hsam, S. L. K. Microsatellite mapping of powdery mildew resistance allele Pm5d from common wheat line IGV1-455. Euphytica 159, 307–313 (2008).
Wang, W. et al. Characterization of the powdery mildew resistance gene in wheat breeding line KN0816 and its evaluation in marker-assisted selection. Plant Dis. 105, 4042–4050 (2021).
Li, H. et al. A spontaneous wheat-Aegilops longissima translocation carrying Pm66 confers resistance to powdery mildew. Theor. Appl. Genet. 133, 1149–1159 (2020).
Wang, Y. et al. Mapping stripe rust resistance gene YrZH22 in Chinese wheat cultivar Zhoumai 22 by bulked segregant RNA-Seq (BSR-Seq) and comparative genomics analyses. Theor. Appl. Genet. 130, 2191–2201 (2017).
Yahiaoui, N., Srichumpa, P., Dudler, R. & Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 37, 528–538 (2004).
Hurni, S. et al. Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76, 957–969 (2013).
Sánchez-Martín, J. et al. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17, 221 (2016).
Zou, S., Wang, H., Li, Y., Kong, Z. & Tang, D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. N. Phytol. 218, 298–309 (2018).
Singh, S. P. et al. Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 98, 249–260 (2018).
Xing, L. et al. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 11, 874–878 (2018).
He, H. et al. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 11, 879–882 (2018).
Li, M. et al. A CNL protein in wild emmer wheat confers powdery mildew resistance. N. Phytol. 228, 1027–1037 (2020).
Xie, J. et al. A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. N. Phytol. 228, 1011–1026 (2020).
Hewitt, T. et al. A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. N. Phytol. 229, 2812–2826 (2021).
Wu, Q. et al. Functional characterization of powdery mildew resistance gene MlIW172, a new Pm60 allele and its allelic variation in wild emmer wheat. J. Genet Genom. 49, 787–795 (2022).
Zhu, S. et al. Orthologous genes Pm12 and Pm21 from two wild relatives of wheat show evolutionary conservation but divergent powdery mildew resistance. Plant Commun. 4, 100472 (2023).
Li, Y. et al. Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69. Plant Commun. 4, 100646 (2023).
Lu, C. et al. Wheat Pm55 alleles exhibit distinct interactions with an inhibitor to cause different powdery mildew resistance. Nat. Commun. 15, 503 (2024).
Gaurav, K. et al. Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 40, 422–431 (2022).
Lu, P. et al. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 11, 680 (2020).
Li, M. et al. A membrane associated tandem kinase from wild emmer wheat confers broad-spectrum resistance to powdery mildew. Nat. Commun. 15, 3124 (2024).
Sánchez-Martín, J. et al. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants 7, 327–341 (2021).
Li, H. et al. Wheat powdery mildew resistance gene Pm13 encodes a mixed lineage kinase domain-like protein. Nat. Commun. 15, 2449 (2024).
Moore, J. W. et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498 (2015).
Krattinger, S. G. et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363 (2009).
Zhu, K. et al. Fine mapping of powdery mildew resistance gene MlWE74 derived from wild emmer wheat (Triticum turgidum ssp. dicoccoides) in an NBS-LRR gene cluster. Theor. Appl. Genet. 135, 1235–1245 (2022).
Friebe, B., Jiang, J., Raupp, W. J., Mcintosh, R. A. & Gill, B. S. Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91, 59–87 (1996).
Liu, W. et al. Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat. Theor. Appl. Genet. 130, 841–848 (2017).
Liu, W. et al. Development and characterization of wheat-Ae. searsii Robertsonian translocations and a recombinant chromosome conferring resistance to stem rust. Theor. Appl. Genet. 122, 1537–1545 (2011).
Dong, Z. et al. Physical mapping of Pm57, a powdery mildew resistance gene derived from Aegilops searsii. Int. J. Mol. Sci. 21, 322 (2020).
Wang, Y. et al. An unusual tandem kinase fusion protein confers leaf rust resistance in wheat. Nat. Genet. 55, 914–920 (2023).
Klymiuk, V., Coaker, G., Fahima, T. & Pozniak, C. J. Tandem protein kinases emerge as new regulators of plant immunity. Mol. Plant. Microbe Interact. 34, 1094–1102 (2021).
Liu, Y., Hou, S. & Chen, S. Kinase fusion proteins: intracellular R-proteins in plant immunity. Trends Plant Sci. 29, 278–282 (2024).
Fahima, T. & Coaker, G. Pathogen perception and deception in plant immunity by kinase fusion proteins. Nat. Genet. 55, 908–909 (2023).
Zang, W. et al. Fine mapping and identification of a candidate gene for the barley Un8 true loose smut resistance gene. Theor. Appl. Genet. 128, 1343–1357 (2015).
Klymiuk, V. et al. Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat. Commun. 9, 3735 (2018).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Lehti-Shiu, M. D. & Shiu, S. H. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 367, 2619–2639 (2012).
Yu, G. et al. Aegilops sharonensis genome-assisted identification of stem rust resistance gene Sr62. Nat. Commun. 13, 1607 (2022).
Zulawski, M., Schulze, G., Braginets, R., Hartmann, S. & Schulze, W. X. The Arabidopsis Kinome: phylogeny and evolutionary insights into functional diversification. BMC Genom. 15, 548 (2014).
Gyawali, Y., Zhang, W., Chao, S., Xu, S. & Cai, X. Delimitation of wheat ph1b deletion and development of ph1b-specific DNA markers. Theor. Appl. Genet. 132, 195–204 (2019).
Brueggeman, R. et al. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. USA 99, 9328–9333 (2002).
Chen, S. et al. Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. N. Phytol. 225, 948–959 (2020).
Arora, S. et al. A wheat kinase and immune receptor form host-specificity barriers against the blast fungus. Nat. Plants 9, 385–392 (2023).
Li, Y., Gou, M., Sun, Q. & Hua, J. Requirement of calcium binding, myristoylation, and protein-protein interaction for the Copine BON1 function in Arabidopsis. J. Biol. Chem. 285, 29884–29891 (2010).
Whittaker, C. A. & Hynes, R. O. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13, 3369–3387 (2002).
Tomsig, J. L., Snyder, S. L. & Creutz, C. E. Identification of targets for calcium signaling through the copine family of proteins. Characterization of a coiled-coil copine-binding motif. J. Biol. Chem. 278, 10048–10054 (2003).
Zou, B. et al. Identification and analysis of copine/BONZAI proteins among evolutionarily diverse plant species. Genome 59, 565–573 (2016).
Zhao, X., Fu, X., Yin, C. & Lu, F. Wheat speciation and adaptation: perspectives from reticulate evolution. Abiotech 2, 386–402 (2021).
Wang, Z. et al. Dispersed emergence and protracted domestication of polyploid wheat uncovered by mosaic ancestral haploblock inference. Nat. Commun. 13, 3891 (2022).
Wang, Z. L. et al. Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89, 457–463 (2005).
Zhu, T. et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 107, 303–314 (2021).
Zhao, Y. et al. Wheat heat shock factor TaHsfA2d contributes to plant responses to phosphate deficiency. Plant Physiol. Biochem. 185, 178–187 (2022).
Zhou, W. et al. TaNAC6s are involved in the basal and broad-spectrum resistance to powdery mildew in wheat. Plant Sci. 277, 218–228 (2018).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Wang, K., Liu, H., Du, L. & Ye, X. Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 15, 614–623 (2017).
Luo, G., Li, B. & Gao, C. Protoplast isolation and transfection in wheat. Methods Mol. Biol. 2464, 131–141 (2022).
Huq, E. & Quail, P. H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450 (2002).
Li, Y. et al. Intracellular reactive oxygen species-aided localized cell death contributing to immune responses against wheat powdery mildew pathogen. Phytopathology 113, 884–892 (2023).
Chen, Y. et al. A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the Triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant 13, 1694–1708 (2020).
Ma, S. et al. WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14, 1965–1968 (2021).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40, D1178–D1186 (2012).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genomics Proteom. Bioinforma. 20, 536–540 (2022).
Zhao, Y. et al. Btr1-A induces grain shattering and affects spike morphology and yield-related traits in wheat. Plant Cell Physiol. 60, 1342–1353 (2019).
Acknowledgements
We are grateful to Prof. Zhongfu Ni and Huiru Peng from China Agricultural University, Prof. Zhiyong Liu from Chinese Academy of Sciences, for their advice and supports during this research. We are also grateful to Prof. Pengtao Ma of Yantai University, Yantai, Shandong, China, for providing Bgt isolates and powdery mildew resistance assays. This research was financially supported by the National Natural Science Foundation of China (31971887 and 32372089 to W.X.L., and 32272070 to H.H.L.), the Scientific and Technological Research Project of Henan Province of China (222103810004 to Y.Z.) and the Key Scientific Research Projects of Higher Education Institutions in Henan Province (23A210020 to Y.Z.) and South Dakota Wheat Commission (3×2030) and USDA AFRI 2022-68013-36439 (WheatCAP) to S.K.S.
Author information
Authors and Affiliations
Contributions
W.X.L., H.H.L. and S.K.S. designed the study. Y.Z., Z.J.D., J.N.M., C.M., X.B.T. and J.Q.H. performed the research. Q.W.L., H.H.B., W.Y., T.L., H.S.G., Z.B.Z., A.Z.C., B.L., H.H.L. and S.K.S. analyzed the data. Y.Z., W.X.L. and S.K.S. wrote the manuscript and all authors contributed to revision and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tzion Fahima, Simon Krattinger, Matthew Moscou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, Y., Dong, Z., Miao, J. et al. Pm57 from Aegilops searsii encodes a tandem kinase protein and confers wheat powdery mildew resistance. Nat Commun 15, 4796 (2024). https://doi.org/10.1038/s41467-024-49257-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-49257-2
- Springer Nature Limited