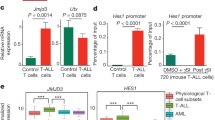
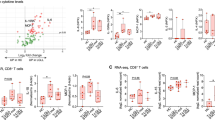
Abstract
T-acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy that comprises the accumulation of malignant T-cells. Despite current therapies, failure to conventional treatments and relapse are frequent in children with T-ALL. It is known that the chemokine CXCL12 modulates leukemia survival and dissemination; however, our understanding of molecular mechanisms used by T-ALL cells to infiltrate and respond to leukemia cells-microenvironment interactions is still vague. In the present study, we showed that CXCL12 promoted H3K9 methylation in cell lines and primary T-ALL cells within minutes. We thus identified that CXCL12-mediated H3K9 methylation affected the global chromatin configuration and the nuclear mechanics of T-ALL cells. Importantly, we characterized changes in the genomic profile of T-ALL cells associated with rapid CXCL12 stimulation. We showed that blocking CXCR4 and protein kinase C (PKC) impaired the H3K9 methylation induced by CXCL12 in T-ALL cells. Finally, blocking H3K9 methyltransferases reduced the efficiency of T-ALL cells to deform their nuclei, migrate across confined spaces, and home to spleen and bone marrow in vivo models. Together, our data show novel functions for CXL12 as a master regulator of nuclear deformability and epigenetic changes in T-ALL cells, and its potential as a promising pharmacological target against T-ALL dissemination.








Similar content being viewed by others
References
Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–43.
Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukemia and lymphoma. Nat Rev Immunol. 2008;8:380–90.
Pui CH, Jeha S. New therapeutic strategies for the treatment of acute lymphoblastic leukaemia. Nat Rev Drug Disco. 2007;6:149–65.
Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. 2001;2:123–128.
Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–88.
de Lourdes Perim A, Amarante MK, Guembarovski RL, de Oliveira CE, Watanabe MA. CXCL12/CXCR4 axis in the pathogenesis of acute lymphoblastic leukemia (ALL): a possible therapeutic target. Cell Mol Life Sci. 2015;72:1715–23.
Pitt LA, Tikhonova AN, Hu H, Trimarchi T, King B, Gong Y, et al. CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance. Cancer Cell. 2015;27:755–68.
Scupoli MT, Donadelli M, Cioffi F, Rossi M, Perbellini O, Malpeli G, et al. Bone marrow stromal cells and the upregulation of interleukin-8 production in human T-cell acute lymphoblastic leukemia through the CXCL12/CXCR4 axis and the NF-kappaB and JNK/AP-1 pathways. Haematologica. 2008;93:524–32.
Cancilla D, Rettig MP, DiPersio JF. Targeting CXCR4 in AML and ALL. Front Oncol. 2020;10:1672.
Hong Z, Wei Z, Xie T, Fu L, Sun J, Zhou F, et al. Targeting chemokines for acute lymphoblastic leukemia therapy. J Hematol Oncol. 2021;14:48.
Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9.
Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64.
Pfeifer CR, Irianto J, Discher DE. Nuclear mechanics and cancer cell migration. Adv Exp Med Biol. 2019;1146:117–30.
Stephens AD, Banigan EJ, Marko JF. Chromatin’s physical properties shape the nucleus and its functions. Curr Opin Cell Biol. 2019;58:76–84.
Schumann K, Lammermann T, Bruckner M, Legler DF, Polleux J, Spatz JP, et al. Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity. 2010;32:703–13.
Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–3.
Kidiyoor GR, Li Q, Bastianello G, Bruhn C, Giovannetti I, Mohamood A, et al. ATR is essential for preservation of cell mechanics and nuclear integrity during interstitial migration. Nat Commun. 2020;11:4828.
Bendall LJ, Baraz R, Juarez J, Shen W, Bradstock KF. Defective p38 mitogen-activated protein kinase signaling impairs chemotaxic but not proliferative responses to stromal-derived factor-1alpha in acute lymphoblastic leukemia. Cancer Res. 2005;65:3290–8.
Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104:15619–24.
Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell. 2017;28:1984–96.
Le Berre M, Aubertin J, Piel M. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol (Camb). 2012;4:1406–14.
Jost TR, Borga C, Radaelli E, Romagnani A, Perruzza L, Omodho L, et al. Role of CXCR4-mediated bone marrow colonization in CNS infiltration by T cell acute lymphoblastic leukemia. J Leukoc Biol. 2016;99:1077–87.
Melo RCC, Longhini AL, Bigarella CL, Baratti MO, Traina F, Favaro P. et al. CXCR7 is highly expressed in acute lymphoblastic leukemia and potentiates CXCR4 response to CXCL12. PLoS One. 2014;9:e85926
Wang JF, Park IW, Groopman JE. Stromal cell-derived factor-1alpha stimulates tyrosine phosphorylation of multiple focal adhesion proteins and induces migration of hematopoietic progenitor cells: roles of phosphoinositide-3 kinase and protein kinase C. Blood. 2000;95:2505–13.
Lai YS, Chen JY, Tsai HJ, Chen TY, Hung WC. The SUV39H1 inhibitor chaetocin induces differentiation and shows synergistic cytotoxicity with other epigenetic drugs in acute myeloid leukemia cells. Blood. Cancer J. 2015;5:e313.
Lu C, Klement JD, Yang D, Albers T, Lebedyeva IO, Waller JL. et al. SUV39H1 regulates human colon carcinoma apoptosis and cell cycle to promote tumor growth. Cancer Lett. 2020;476:87–96.
Irianto J, Xia Y, Pfeifer CR, Greenberg RA, Discher DE. As a nucleus enters a small pore, chromatin stretches and maintains integrity, even with DNA breaks. Biophys J. 2017;112:446–9.
Vargas P, Terriac E, Lennon-Dumenil AM, Piel M. Study of cell migration in microfabricated channels. J Vis Exp. 2014;84:e51099.
Cannon JL, Oruganti SR, Vidrine DW. Molecular regulation of T-ALL cell infiltration into the CNS. Oncotarget. 2017;8:84626–7.
Frishman-Levy L, Izraeli S. Advances in understanding the pathogenesis of CNS acute lymphoblastic leukemia and potential for therapy. Br J Haematol. 2017;176:157–67.
Vadillo E, Dorantes-Acosta E, Pelayo R, Schnoor M. T cell acute lymphoblastic leukemia (T-ALL): New insights into the cellular origins and infiltration mechanisms common and unique among hematologic malignancies. Blood Rev. 2018;32:36–51.
Gomez AM, Martinez C, Gonzalez M, Luque A, Melen GJ, Martínez J, et al. Chemokines and relapses in childhood acute lymphoblastic leukemia: A role in migration and in resistance to antileukemic drugs. Blood Cells Mol Dis. 2015;55:220–7.
Bradstock KF, Makrynikola V, Bianchi A, Shen W, Hewson J, Gottlieb DJ. Effects of the chemokine stromal cell-derived factor-1 on the migration and localization of precursor-B acute lymphoblastic leukemia cells within bone marrow stromal layers. Leukemia. 2000;14:882–8.
Kato I, Niwa A, Heike T, Fujino H, Saito MK, Umeda K, et al. Identifcation of hepatic niche harboring human acute lymphoblastic leukemic cells via the SDF-1/CXCR4 axis. PLoS ONE. 2011;6:e27042.
Kuang SQ, Fang Z, Zweidler-McKay PA, Yang H, Wei Y, Gonzalez-Cervantes EA, et al. Epigenetic inactivation of Notch-Hes pathway in human B-cell acute lymphoblastic leukemia. PLoS One. 2013;8:e61807.
Crazzolara R, Kreczy A, Mann G, Heitger A, Eibl G, Fink FM. et al. High expression of the chemokine receptor CXCR4 predicts extramedullary organ infiltration in childhood acute lymphoblastic leukaemia. Br J Haematol. 2001;115:545–53.
Bao Y, Wang Z, Liu B, Lu X, Xiong Y, Shi J, et al. A feed-forward loop between nuclear translocation of CXCR4 and HIF-1alpha promotes renal cell carcinoma metastasis. Oncogene. 2019;38:881–95.
Alsadeq A, Fedders H, Vokuhl C, Belau NM, Zimmermann M, Wirbelauer T, et al. The role of ZAP70 kinase in acute lymphoblastic leukemia infiltration into the central nervous system. Haematologica. 2017;102:346–55.
Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood. 2009;113:6085–93.
Song ZY, Wang F, Cui SX, Gao ZH. Qu XJ.CXCR7/CXCR4 heterodimer-induced histone demethylation: a new mechanism of colorectal tumorigenesis. Oncogene. 2019;38:1560–75.
Altman A, Kong KF. Protein kinase C enzymes in the hematopoietic and immune systems. Annu Rev Immunol. 2016;34:511–38.
Petit I, Goichberg P, Spiegel A, Peled A, Brodie C, Seger R. et al. Atypical PKC-zeta regulates SDF-1-mediated migration and development of human CD34+ progenitor cells. J Clin Invest. 2005;115:168–76.
Wei SY, Lin TE, Wang WL, Lee PL, Tsai MC, Chiu JJ. Protein kinase C-δ and -β coordinate flow-induced directionality and deformation of migratory human blood T-lymphocytes. J Mol Cell Biol. 2014;6:458–72.
Guinamard R, Signoret N, Ishiai M, Marsh M, Kurosaki T, Ravetch JV. B cell antigen receptor engagement inhibits stromal cell-derived factor (SDF)-1alpha chemotaxis and promotes protein kinase C (PKC)-induced internalization of CXCR4. J Exp Med. 1999;189:1461–6.
Velázquez-Avila M, Balandrán JC, Ramírez-Ramírez D, Velázquez-Avila M, Sandoval A, Felipe-López A, et al. High cortactin expression in B-cell acute lymphoblastic leukemia is associated with increased transendothelial migration and bone marrow relapse. Leukemia. 2019;33:1337–48.
Poli A, Ratti S, Finelli C, Mongiorgi S, Clissa C, Lonetti A, et al. Nuclear translocation of PKC-alpha is associated with cell cycle arrest and erythroid differentiation in myelodysplastic syndromes (MDSs). FASEB J. 2018;32:681–92.
Edens LJ, Dilsaver MR, Levy DL. PKC-mediated phosphorylation of nuclear lamins at a single serine residue regulates interphase nuclear size in Xenopus and mammalian cells. Mol Biol Cell. 2017;28:1389–99.
Esteller M. Epigenetics in cancer. N. Engl J Med. 2008;358:1148–59.
Almamun M, Levinson BT, van Swaay AC, Johnson NT, McKay SD, Arthur GL, et al. Integrated methylome and transcriptome analysis reveals novel regulatory elements in pediatric acute lymphoblastic leukemia. Epigenetics. 2015;10:882–90.
Konopleva MY, Jordan CT. Leukemia stem cells and microenvironment: biology and therapeutic targeting. J Clin Oncol. 2011;29:591–9.
Passaro D, Irigoyen M, Catherinet C, Gachet S, Da Costa C, Lasgi C, et al. CXCR4 is required for Leukemia-Initiating Cell activity in T cell acute lymphoblastic leukemia. Cancer Cell. 2015;27:769–79.
Nava MM, Miroshnikova YA, Biggs LC, Whitefield DB, Metge F, Boucas J, et al. Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell. 2020;181:800–17.
Makhija E, Jokhun DS, Shivashankar GV. Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc Natl Acad Sci USA. 2016;113:E32–40.
Pfeifer CR, Xia Y, Zhu K, Liu D, Irianto J, García VMM, et al. Constricted migration increases DNA damage and independently represses cell cycle. Mol Biol Cell. 2018;29:1948–62.
Zhang X, Cook PC, Zindy E, Williams CJ, Jowitt TA, Streuli CH, et al. Integrin alpha4beta1 controls G9a activity that regulates epigenetic changes and nuclear properties required for lymphocyte migration. Nucleic Acids Res. 2016;44:3031–44.
Gerlitz G, Bustin M. Efficient cell migration requires global chromatin condensation. J Cell Sci. 2010;123:2207–17.
Madrazo E, Ruano D, Abad L, Alonso-Gómez E, Sánchez-Valdepeñas C, González-Murillo A. et al. G9a Correlates with VLA-4 integrin and influences the migration of childhood acute lymphoblastic leukemia cells. Cancers (Basel). 2018;10:325
Sison EA, Magoon D, Li L, Annesley CE, Romagnoli B, Douglas GJ, et al. POL5551, a novel and potent CXCR4 antagonist, enhances sensitivity to chemotherapy in pediatric ALL. Oncotarget. 2015;6:30902–18.
Randhawa S, Cho BS, Ghosh D, Sivina M, Koehrer S, Müschen M, et al. Effects of pharmacological and genetic disruption of CXCR4 chemokine receptor function in B-cell acute lymphoblastic leukaemia. Br J Haematol. 2016;174:425–36.
Barbieri F, Bajetto A, Thellung S, Würth R, Florio T. Drug design strategies focusing on the CXCR4/CXCR7/CXCL12 pathway in leukemia and lymphoma. Expert Opin Drug Disco. 2016;11:1093–109.
Redondo-Muñoz J, García-Pardo A, Teixidó J. Molecular players in hematologic tumor cell trafficking. Front Immunol. 2019;10:156.
San José-Enériz E, Agirre X, Rabal O, Vilas-Zornoza A, Sanchez-Arias JA, Miranda E, et al. Discovery of first-in-class reversible dual small molecule inhibitors against G9a and DNMTs in hematological malignancies. Nat Commun. 2017;8:15424.
Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122:1469–86.
Yokoyama M, Chiba T, Zen Y, Oshima M, Kusakabe Y, Noguchi Y, et al. Histone lysine methyltransferase G9a is a novel epigenetic target for the treatment of hepatocellular carcinoma. Oncotarget. 2017;8:21315–26.
Isham CR, Tibodeau JD, Jin W, Xu R, Timm MM, Bible KC. Chaetocin: a promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress. Blood. 2007;109:2579–88.
Jung HJ, Seo I, Casciello F, Jacquelin S, Lane SW, Suh SI, et al. The anticancer effect of chaetocin is enhanced by inhibition of autophagy. Cell Death Dis. 2016;7:e2098.
Acknowledgements
The authors thank the Microscopy Unit and the Flow Cytometry Core Unit of Instituto de Investigación Biosanitaria Gregorio Marañón (IiSGM) for assistance with confocal, videomicroscopy and flow cytometry analyses. The UCM-Genomic CAI Unit for their assistance with the microarray experiments. We thank Dr. Ignacio Casal, Joaquín Teixidó, Alicia García-Arroyo and María Montoya for insightful and critical comments. This research was supported by a FPI Scholarship 2018 (Ministerio de Ciencia e Innovacion/MICINN, Agencia Estatal de Investigacion/AEI y Fondo Europeo de Desarrollo Regional/FEDER) to R.G.N.; an undergraduate fellowship “Beca de Introducción a la Investigación” (Asociación Española Contra el Cáncer) to C.O.P.; and by grants from Asociación Pablo Ugarte to M.R., Gilead Sciences International Scholar in Hematology/Oncology (Gilead), 2020 Leonardo Grant for Researchers and Cultural Creators (BBVA Foundation) and SAF2017-86327-R (Ministerio de Ciencia e Innovacion/MICINN, Agencia Estatal de Investigacion/AEI y Fondo Europeo de Desarrollo Regional/FEDER) to J.R.M.
Author information
Authors and Affiliations
Contributions
EM carried out biochemical and imaging experiments and contributed to the interpretation of biological data. RGN performed biochemical and in vivo studies. COP contributed to biochemical and imaging experiments. AGM provided clinically annotated multiple ALL samples and contributed to in vivo experiments and the interpretation of clinical data. MGL carried out the bioinformatics analysis of ChIP-seq and microarray data samples and contributed to the interpretation of data. MR designed part of the studies, provided ALL samples, and contributed to the interpretation of clinical data. JRM designed the studies, supervised the project, and wrote the manuscript with the input of all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Madrazo, E., González-Novo, R., Ortiz-Placín, C. et al. Fast H3K9 methylation promoted by CXCL12 contributes to nuclear changes and invasiveness of T-acute lymphoblastic leukemia cells. Oncogene 41, 1324–1336 (2022). https://doi.org/10.1038/s41388-021-02168-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02168-8
- Springer Nature Limited
This article is cited by
-
Mechanical stress confers nuclear and functional changes in derived leukemia cells from persistent confined migration
Cellular and Molecular Life Sciences (2023)
-
Current methods for studying metastatic potential of tumor cells
Cancer Cell International (2022)