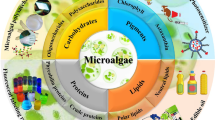
Abstract
In modern human civilization, the demand for lipids has become upswing for several purposes: nutritional supplements, the production of foods, surfactants, lubricants, and biofuels. With the gradual rise in population, shortage, and deterioration of arable land due to anthropogenic activities, traditional lipids production methods alone cannot conciliate future demand. Various microbial genera related to algae, bacteria, fungi, and yeast can synthesize and accumulate lipids in their bodies. Currently, microbial lipids have emerged as a sustainable successor of plant-derived lipids. However, the commercial scale production of microbial lipids faces some problems such as inadequate level of lipid accumulation in the microbial cells, lipid extraction, and operational cost associated with microbial cultivation. Thus, there is an urgent need to construct oleaginous microbes with modified, preferable features. With the modern biotechnological tools, the insights of the complex microbial genetic makeup and metabolic pathways become explored, allowing various genetic engineering (GE) and metabolic engineering (ME) approaches to develop microbes of desired lipid production abilities with the required lipid profile and physiological qualities. The current review mainly deals with the basic lipid metabolic pathways run in the various groups of microbes, properties of lipids they synthesized, state-of-the-art GE and ME techniques useful for the lipid overproduction in oleaginous microbes, and explain challenging futuristic developmental directions.


Similar content being viewed by others
Data availability
Not applicable.
References
Ajjawi I, Verruto J, Aqui M, Soriaga LB, Coppersmith J, Kwok K, Moellering ER. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat Biotechnol. 2017;35(7):647–52.
Alvarez HM. Triacylglycerol and wax ester-accumulating machinery in prokaryotes. Biochimie. 2016;120:28–39.
Anantayanon J, Jeennor S, Panchanawaporn S, Chutrakul C, Laoteng K. Significance of two intracellular triacylglycerol lipases of Aspergillus oryzae in lipid mobilization: a perspective in industrial implication for microbial lipid production. Gene. 2021;793:145745. https://doi.org/10.1016/j.gene.2021.145745.
Angerbauer C, Siebenhofer M, Mittelbach M, Guebitz GM. Conversion of sewage sludge into lipids by Lipomyces starkeyi for biodiesel production. Bioresour Technol. 2008;99(8):3051–6.
Aratboni HA, Rafiei N, Garcia-Granados R, Alemzadeh A, Morones-Ramírez JR. Biomass and lipid induction strategies in microalgae for biofuel production and other applications. Microb Cell Fact. 2019;18(1):1–17.
Arguelles ED, Laurena AC, Monsalud RG, Martinez-Goss MR. Fatty acid profile and fuel- derived physico-chemical properties of biodiesel obtained from an indigenous green microalga, Desmodesmus sp. (I-AU1), as potential source of renewable lipid and high-quality biodiesel. J Appl Phycol. 2018;30(1):411–9.
Awad D, Younes S, Glemser M, Wagner FM, Schenk G, Mehlmer N, Brueck T. Towards high-throughput optimization of microbial lipid production: from strain development to process monitoring. Sustain Energy Fuels. 2020;4(12):5958–69.
Bartley ML, Boeing WJ, Corcoran AA, Holguin FO, Schaub T. Effects of salinity on growth and lipid accumulation of biofuel microalga Nannochloropsis salina and invading organisms. Biomass Bioenerg. 2013;54:83–8.
Battah M, El-Ayoty Y, Abomohra AEF, Abd El-Ghany S, Esmael A. Effect of Mn2+, Co2+ and H2O2 on biomass and lipids of the green microalga Chlorella vulgaris as a potential candidate for biodiesel production. Annals Microbiol. 2015;65(1):155–62.
Berg JM, Tymoczko JL, Stryer L. Biochemistry. Freeman Macmillan; 2015.
Bharathiraja B, Sridharan S, Sowmya V, Yuvaraj D, Praveenkumar R. Microbial oil–a plausible alternate resource for food and fuel application. Bioresour Technol. 2017;233:423–32.
Cao S, Zhou X, Jin W, Wang F, Tu R, Han S, Ma F. Improving of lipid productivity of the oleaginous microalgae Chlorella pyrenoidosa via atmospheric and room temperature plasma (ARTP). Bioresour Technol. 2017;244:1400–6.
Carsanba E, Papanikolaou S, Fickers P, Erten H. Lipids by Yarrowia lipolytica strains cultivated on glucose in batch cultures. Microorganisms. 2020;8(7):1054.
Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallström BM, Nielsen J. Altered sterol composition renders yeast thermotolerant. Science. 2014;346(6205):75–8.
Chao YANG, Guo ZB, Du ZM, Yang HY, Bi YJ, Wang GQ, Tan YF. Cellular fatty acids as chemical markers for differentiation of Acinetobacter baumannii and Acinetobacter calcoaceticus. Biomed Environ Sci. 2012;25(6):711–7.
Chattopadhyay A, Mitra M, Maiti MK. Recent advances in lipid metabolic engineering of oleaginous yeasts. Biotechnol Adv. 2021. https://doi.org/10.1016/j.biotechadv.2021.107722.
Chen CY, Lee MH, Dong CD, Leong YK, Chang JS. Enhanced production of microalgal lipids using a heterotrophic marine microalga Thraustochytrium sp. BM2. Biochem Eng J. 2020;154:107429.
Chen H, Li M, Liu C, Zhang H, Xian M, Liu H. Enhancement of the catalytic activity of Isopentenyl diphosphate isomerase (IDI) from Saccharomyces cerevisiae through random and site-directed mutagenesis. Microbial Cell Fact. 2018;17(1):1–14.
Chen HH, Jiang JG. Lipid accumulation mechanisms in auto-and heterotrophic microalgae. J Agric Food Chem. 2017;65(37):8099–110.
Chen L, Lee J, Chen WN. The use of metabolic engineering to produce fatty acid-derived biofuel and chemicals in Saccharomyces cerevisiae: a review. AIMS Bioeng. 2016;3(4):468–92.
Choi KR, Jang WD, Yang D, Cho JS, Park D, Lee SY. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019;37(8):817–37.
Choi YJ, Lee SY. Microbial production of short-chain alkanes. Nature. 2013;502(7472):571–4.
Chowdury KH, Nahar N, Deb UK. The growth factors involved in microalgae cultivation for biofuel production: a review. Comput Water Energy Environ Eng. 2020;9(4):185–215.
Daboussi F, Leduc S, Marechal A, Dubois G, Guyot V, Perez-Michaut C, Duchateau P. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat Commun. 2014;5(1):1–7.
Danouche M, El Ghachtouli N, El Arroussi H. Phycoremediation mechanisms of heavy metals using living green microalgae: physicochemical and molecular approaches for enhancing selectivity and removal capacity. Heliyon. 2021. https://doi.org/10.1016/j.heliyon.2021.e07609.
Darwish R, Gedi MA, Akepach P, Assaye H, Zaky AS, Gray DA. Chlamydomonas reinhardtii: is a potential food supplement with the capacity to outperform Chlorella and Spirulina. Appl Sci. 2020;10(19):6736.
Daskalaki A, Perdikouli N, Aggeli D, Aggelis G. Laboratory evolution strategies for improving lipid accumulation in Yarrowia lipolytica. Appl Microbiol Biotechnol. 2019;103(20):8585–96.
de Jesús-Campos D, López-Elías JA, Medina-Juarez LÁ, Carvallo-Ruiz G, Fimbres-Olivarria D, Hayano-Kanashiro C. Chemical composition, fatty acid profile and molecular changes derived from nitrogen stress in the diatom Chaetoceros muelleri. Aquac Rep. 2020;16: 100281.
Deng X, Fan X, Li P, Fei X. A photoperiod-regulating gene CONSTANS is correlated to lipid biosynthesis in Chlamydomonas reinhardtii. BioMed Res Int. 2015. https://doi.org/10.1155/2015/715020.
Dourou M, Aggeli D, Papanikolaou S, Aggelis G. Critical steps in carbon metabolism affecting lipid accumulation and their regulation in oleaginous microorganisms. Appl Microbiol Biotechnol. 2018;102(6):2509–23.
Dulermo T, Nicaud JM. Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab Eng. 2011;13(5):482–91.
El Maghraby DM, Fakhry EM. Lipid content and fatty acid composition of Mediterranean macro-algae as dynamic factors for biodiesel production. Oceanologia. 2015;57(1):86–92.
El-Kantar S, Khelfa A, Vorobiev E, Koubaa M. Strategies for increasing lipid accumulation and recovery from Y. lipolytica: a review. OCL. 2021;28:51. https://doi.org/10.1051/ocl/2021038.
Elnar AG, Kim MG, Lee JE, Han RH, Yoon SH, Lee GY, Kim GB. Acinetobacter pullorum sp. nov., Isolated from chicken meat. J Microbiol Biotechnol. 2020;30(4):526–32.
El-Sheekh M, Abomohra AEF, Abd El-Azim M, Abou-Shanab R. Effect of temperature on growth and fatty acids profile of the biodiesel producing microalga Scenedesmus acutus. BASE. 2017. https://doi.org/10.25518/1780-4507.15291.
Fakas S. Lipid biosynthesis in yeasts: a comparison of the lipid biosynthetic pathway between the model nonoleaginous yeast Saccharomyces cerevisiae and the model oleaginous yeast Yarrowia lipolytica. Eng Life Sci. 2017;17(3):292–302.
Garay LA, Boundy-Mills KL, German JB. Accumulation of high-value lipids in single-cell microorganisms: a mechanistic approach and future perspectives. J Agric Food Chem. 2014;62(13):2709–27.
Gardeli C, Athenaki M, Xenopoulos E, Mallouchos AKAA, Koutinas AA, Aggelis G, Papanikolaou S. Lipid production and characterization by Mortierella (Umbelopsis) isabellina cultivated on lignocellulosic sugars. J Appl Microbiol. 2017;123(6):1461–77.
Gaur JP, Rai LC. Heavy metal tolerance in algae. In: Gaur JP, Rai LC, editors. Algal adaptation to environmental stresses. Heidelberg: Springer; 2001. p. 363–88.
Ghasemi M, Atakishiyeva Y, Asadi A. The changes in lipid composition of Pythium irregulare LX oomycetes at a stressful situation created with crude oil. Mol Biol Res Commun. 2012;1(1):33–8.
Ghogare R, Chen S, Xiong X. Metabolic engineering of oleaginous yeast Yarrowia lipolytica for overproduction of fatty acids. Front Microbiol. 2020;11:1717. https://doi.org/10.3389/fmicb.2020.01717.
Goswami L, Namboodiri MT, Kumar RV, Pakshirajan K, Pugazhenthi G. Biodiesel production potential of oleaginous Rhodococcus opacus grown on biomass gasification wastewater. Renew Energ. 2017;105:400–6.
Guha TK, Wai A, Hausner G. Programmable genome editing tools and their regulation for efficient genome engineering. Comput Struct Biotechnol J. 2017;15:146–60.
Gupta N, Manikandan NA, Pakshirajan K. Real-time lipid production and dairy wastewater treatment using Rhodococcus opacus in a bioreactor under fed-batch, continuous and continuous cell recycling modes for potential biodiesel application. Biofuels. 2018;9(2):239–45.
Guschina IA, Harwood JL. Algal lipids and effect of the environment on their biochemistry. In: Kainz M, Brett MT, Arts MT, editors. Lipids in aquatic ecosystems. New York: Springer; 2009. p. 1–24.
Guschina IA, Harwood JL. Algal lipids and their metabolism. In: Borowitzka MA, Moheimani NR, editors. Algae for biofuels and energy. Dordrecht: Springer; 2013. p. 17–36.
Hamilton ML, Haslam RP, Napier JA, Sayanova O. Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metabolic Eng. 2014;22:3–9.
Hammerschlag A. Bacteriologisch-Chemische Untersuchungen der Tuberkelbacillen. Monatsh Chem. 1889;10:9–18. https://doi.org/10.1007/BF01516412.
Hao G, Barker GC. Fatty acid secretion by the white-rot fungus, Trametes versicolor. J Ind Microbiol Biotechnol. 2022;49(1):kuab083. https://doi.org/10.1093/jimb/kuab083.
Hashem AH, Abu-Elreesh G, El-Sheikh HH, Suleiman WB. Isolation, identification, and statistical optimization of a psychrotolerant Mucor racemosus for sustainable lipid production. Biomass Convers Biorefinery. 2023;13(4):3415–26.
Heil CS, Wehrheim SS, Paithankar KS, Grininger M. Fatty acid biosynthesis: chain-length regulation and control. Chem BioChem. 2019;20(18):2298–321.
Hernández MA, Alvarez HM. Increasing lipid production using an NADP+-dependent malic enzyme from Rhodococcus jostii. Microbiology. 2019;165(1):4–14.
Hwangbo M, Chu KH. Recent advances in production and extraction of bacterial lipids for biofuel production. Sci Total Environ. 2020;734: 139420.
Ivancic SM, Santek B, Beluhan S. Production of microbial lipids from lignocellulosic biomass. Adv Biofuels Bioenergy. 2018. https://doi.org/10.5772/intechopen.74013.
Jang HD, Lin YY, Yang SS. Effect of culture media and conditions on polyunsaturated fatty acids production by Mortierella alpina. Bioresour Technol. 2005;96(15):1633–44.
Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Mergeay M. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One. 2010;5(5): e10433.
Jay MI, Kawaroe M, Effendi H. Lipid and fatty acid composition microalgae Chlorella vulgaris using photobioreactor and open pond. In IOP Conference Series: Earth and Environmental Science. IOP Publishing. https://doi.org/10.1088/1755-1315/141/1/012015
Jeon S, Koh HG, Cho JM, Kang NK, Chang YK. Enhancement of lipid production in Nannochloropsis salina by overexpression of endogenous NADP-dependent malic enzyme. Algal Res. 2021;54:102218. https://doi.org/10.1016/j.algal.2021.102218.
Jiang M, Guo B, Wan X, Gong Y, Zhang Y, Hu C. Isolation and characterization of the diatom Phaeodactylum Δ5-elongase gene for transgenic LC-PUFA production in Pichia pastoris. Mar Drugs. 2014;12(3):1317–34.
Jin M, Slininger PJ, Dien BS, Waghmode S, Moser BR, Orjuela A, Balan V. Microbial lipid-based lignocellulosic biorefinery: feasibility and challenges. Trends Biotechnol. 2015;33(1):43–54.
Jones AD, Boundy-Mills KL, Barla GF, Kumar S, Ubanwa B, Balan V. Microbial lipid alternatives to plant lipids. In: Balan V, editor. Microbial lipid production. New York: Humana; 2019. p. 1–32.
Jung JH, Sirisuk P, Ra CH, Kim JM, Jeong GT, Kim SK. Effects of green LED light and three stresses on biomass and lipid accumulation with two-phase culture of microalgae. Process Biochem. 2019;77:93–9.
Kan G, Shi C, Wang X, Xie Q, Wang M, Wang X, Miao J. Acclimatory responses to high- salt stress in Chlamydomonas (Chlorophyta, Chlorophyceae) from Antarctica. Acta Oceanologica Sin. 2012;31(1):116–24.
Kang NK, Kim EK, Kim YU, Lee B, Jeong WJ, Jeong BR, Chang YK. Increased lipid production by heterologous expression of AtWRI1 transcription factor in Nannochloropsis salina. Biotechnol Biofuels. 2017;10(1):1–14.
Kanti A, Sukara E, Latifah K, Sukarno N, Boundy-Mills K. Indonesian oleaginous yeasts isolated from Piper betle and P. nigrum. Mycosphere. 2013;4(3):363–454.
Karpagam R, Preeti R, Ashokkumar B, Varalakshmi P. Enhancement of lipid production and fatty acid profiling in Chlamydomonas reinhardtii, CC1010 for biodiesel production. Ecotoxicol Environ Saf. 2015;121:253–7.
Kim BG, Park BG, Kim J, Kim EJ, Kim Y, Kim J, Kim JY. Application of random mutagenesis and synthetic FadR promoter for de novo production of ω-hydroxy fatty acid in Yarrowia lipolytica. Front Bioeng Biotechnol. 2021. https://doi.org/10.3389/fbioe.2021.624838.
Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–45.
Kohlwein SD. Analyzing and understanding lipids of yeast: a challenging endeavor. Cold Spring Harb Protoc. 2017. https://doi.org/10.1101/pdb.top078956.
Koreti D, Kosre A, Jadhav SK, Chandrawanshi NK. A comprehensive review on oleaginous bacteria: an alternative source for biodiesel production. Bioresour Bioprocess. 2022;9(1):1–19.
Kot AM, Błażejak S, Kurcz A, Gientka I, Kieliszek M. Rhodotorula glutinis-potential source of lipids, carotenoids, and enzymes for use in industries. Appl Microbiol Biotechnol. 2016;100(14):6103–17.
Kumar S, Gupta N, Pakshirajan K. Simultaneous lipid production and dairy wastewater treatment using Rhodococcus opacus in a batch bioreactor for potential biodiesel application. J Environ Chem Eng. 2015;3(3):1630–6.
Kumari P, Kumar M, Reddy CRK, Jha B. Algal lipids, fatty acids and sterols. In: Domínguez M, editor. Functional ingredients from algae for foods and nutraceuticals. Woodhead Publishing; 2013. p. 87–134.
Lamers D, Visscher B, Weusthuis RA, Francke C, Wijffels RH, Lokman C. Overexpression of delta-12 desaturase in the yeast Schwanniomyces occidentalis enhances the production of linoleic acid. Bioresour Technol. 2019;289: 121672.
Ledesma-Amaro R. Microbial oils: a customizable feedstock through metabolic engineering. Eur J Lipid Sci Technol. 2015;117(2):141–4.
Lee JE, Vadlani PV, Min D. Sustainable production of microbial lipids from lignocellulosic biomass using oleaginous yeast cultures. J Sustain Bioenergy Syst. 2017;7(1):36–50.
Li DW, Balamurugan S, Yang YF, Zheng JW, Huang D, Zou LG, Li HY. Transcriptional regulation of microalgae for concurrent lipid overproduction and secretion. Sci Advan. 2019;5(1):eaau3795.
Li Z, Meng T, Ling X, Li J, Zheng C, Shi Y, He N. Overexpression of malonyl-CoA: ACP transacylase in Schizochytrium sp. to improve polyunsaturated fatty acid production. J Agric Food Chem. 2018;66(21):5382–91.
Li Z, Sun H, Mo X, Li XXB, Tian P. Overexpression of malic enzyme (ME) of Mucor circinelloides improved lipid accumulation in engineered Rhodotorula glutinis. Appl Microbiol Biotechnol. 2013;97(11):4927–36.
Lin WR, Ng IS. Development of CRISPR/Cas9 system in Chlorella vulgaris FSP-E to enhance lipid accumulation. Enzyme Microbial Technol. 2020;133:109458.
Lin H, Castro NM, Bennett GN, San KY. Acetyl-CoA synthetase overexpression in Escherichia coli demonstrates more efficient acetate assimilation and lower acetate accumulation: a potential tool in metabolic engineering. Appl Microbiol Biotechnol. 2006;71(6):870–4.
Liu X, Zhang D, Zhang J, Chen Y, Liu X, Fan C, Hu Z. Overexpression of the transcription factor AtLEC1 significantly improved the lipid content of Chlorella ellipsoidea. Front Bioeng Biotechnol. 2021;9:113.
Lopez-Lara IM, Geiger O. Bacterial lipid diversity. Biochem Biophys Acta Mol Cell Biol Lipids. 2017;1862(11):1287–99.
Lu X, Vora H, Khosla C. Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab Eng. 2008;10(6):333–9.
Magdouli S, Brar SK, Blais JF. Co-culture for lipid production: advances and challenges. Biomass Bioenergy. 2016;92:20–30.
Mamatha SS, Ravi R, Venkateswaran G. Medium optimization of gamma linolenic acid production in Mucor rouxii CFR-G15 using RSM. Food Bioproc Technol. 2008;1(4):405–9.
Marella ER, Holkenbrink C, Siewers V, Borodina I. Engineering microbial fatty acid metabolism for biofuels and biochemicals. Curr Opin Biotechnol. 2018;50:39–46.
Markham KA, Alper HS. Engineering Yarrowia lipolytica for the production of cyclopropanated fatty acids. J Ind Microbiol Biotechnol. 2018;45(10):881–8.
Martin MA. First generation biofuels compete. New Biotechnol. 2010;27(5):596–608.
Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802.
Mata TM, Almeidab R, Caetanoa NS. Effect of the culture nutrients on the biomass and lipid productivities of microalgae Dunaliella tertiolecta. Chem Eng. 2013;32:973.
Matsakas L, Giannakou M, Vörös D. Effect of synthetic and natural media on lipid production from Fusarium oxysporum. Electron J Biotechnol. 2017;30:95–102.
Matsakas L, Sterioti AA, Rova U, Christakopoulos P. Use of dried sweet sorghum for the efficient production of lipids by the yeast Lipomyces starkeyi CBS 1807. Ind Crops Prod. 2014;62:367–72.
Mavrommati M, Daskalaki A, Papanikolaou S, Aggelis G. Adaptive laboratory evolution principles and applications in industrial biotechnology. Biotechnol Advan. 2021. https://doi.org/10.1016/j.biotechadv.2021.107795.
Meng X, Yang J, Cao Y, Li L, Jiang X, Xu X, Zhang Y. Increasing fatty acid production in E. coli by simulating the lipid accumulation of oleaginous microorganisms. J Ind Microbiol Biotechnol. 2011;38(8):919–25.
Metsoviti MN, Papapolymerou G, Karapanagiotidis IT, Katsoulas N. Effect of light intensity and quality on growth rate and composition of Chlorella vulgaris. Plants. 2020;9(1):31.
Moi IM, Leow ATC, Ali MSM, Rahman RNZRA, Salleh AB, Sabri S. Polyunsaturated fatty acids in marine bacteria and strategies to enhance their production. Appl Microbiol Biotechnol. 2018;102(14):5811–26.
Mondal S, Halder SK, Mondal KC. Tailoring in fungi for next generation cellulase production with special reference to CRISPR/CAS system. Syst Microbiol Biomanuf. 2021. https://doi.org/10.1007/s43393-021-00045-9.
Munoz CF, Sturme MH, D'Adamo S, Weusthuis RA, Wijffels RH. Stable transformation of the green algae Acutodesmus obliquus and Neochloris oleoabundans based on E. coli conjugation. Algal Res. 2019;39:101453.
Muro E, Atilla-Gokcumen GE, Eggert US. Lipids in cell biology: how can we understand them better? Mol Biol Cell. 2014;25(12):1819–23.
Mussgnug JH, Thomas-Hall S, Rupprecht J, Foo A, Klassen V, McDowall A, Hankamer B. Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnol J. 2007;5(6):802–14.
Nelson DL, Cox MM. Lehninger principles of biochemistry. Macmillan; 2017.
Nicolau R, Galera-Cunha A, Lucas Y. Transfer of nutrients and labile metals from the continent to the sea by a small Mediterranean river. Chemosphere. 2006;63(3):469–76.
Nielsen J, Keasling JD. Engineering cellular metabolism. Cell. 2016;164(6):1185–97.
Nouri H, Moghimi H, Rad MN, Ostovar M, Mehr SSF, Ghanaatian F, Talebi AF. Enhanced growth and lipid production in oleaginous fungus, Sarocladium kiliense ADH17: study on fatty acid profiling and prediction of biodiesel properties. Renew Energ. 2019;135:10–20.
Núñez-Cardona MT. Influence of culture conditions on the fatty acids composition of green and purple photosynthetic sulphur bacteria. In: Guo X, editor. Advances in gas chromatography. InTech; 2014.
Ochsenreither K, Glück C, Stressler T, Fischer L, Syldatk C. Production strategies and applications of microbial single cell oils. Front Microbiol. 2016;7:1539.
Ortiz Montoya EY, Casazza AA, Aliakbarian B, Perego P, Converti A, de Carvalho JCM. Production of Chlorella vulgaris as a source of essential fatty acids in a tubularphotobioreactor continuously fed with air enriched with CO2 at different concentrations. Biotechnol Prog. 2014;30(4):916–22.
Ottenheim C, Nawrath M, Wu JC. Microbial mutagenesis by atmospheric and room- temperature plasma (ARTP): the latest development. Bioresourc Bioprocess. 2018;5(1):1–14.
Ouyang LL, Li H, Yan XJ, Xu JL, Zhou ZG. Site-directed mutagenesis from Arg195 to His of a microalgal putatively chloroplastidial glycerol-3-phosphate acyltransferase causes an increase in phospholipid levels in yeast. Front Plant Sci. 2016;7:286.
Papanikolaou S, Aggelis G. Lipids of oleaginous yeasts. Part I: biochemistry of single cell oil production. Eur J Lipid Sci Technol. 2011;113(8):1031–51.
Patel A, Antonopoulou I, Enman J, Rova U, Christakopoulos P, Matsakas L. Lipids detection and quantification in oleaginous microorganisms: an overview of the current state of the art. BMC Chemical Eng. 2019;1(1):1–25.
Patel A, Karageorgou D, Rova E, Katapodis P, Rova U, Christakopoulos P, Matsakas L. An overview of potential oleaginous microorganisms and their role in biodiesel and omega-3 fatty acid-based industries. Microorganisms. 2020;8(3):434.
Patel A, Mikes F, Bühler S, Matsakas L. Valorization of brewers’ spent grain for the production of lipids by oleaginous yeast. Molecules. 2018;23(12):3052.
Patel A, Sartaj K, Pruthi PA, Pruthi V, Matsakas L. Utilization of clarified butter sediment waste as a feedstock for cost-effective production of biodiesel. Foods. 2019;8(7):234.
Patel A, Sindhu DK, Arora N, Singh RP, Pruthi V, Pruthi PA. Biodiesel production from non-edible lignocellulosic biomass of Cassia fistula L. fruit pulp using oleaginous yeast Rhodosporidium kratochvilovae HIMPA1. Bioresour Technol. 2015;197:91–8.
Pathak VM. Review on the current status of polymer degradation: a microbial approach. Bioresour Bioprocess. 2017;4(1):1–31.
Patnaik R, Mallick N. Microalgal biodiesel production: realizing the sustainability index. Front Bioeng Biotechnol. 2021;9:620777. https://doi.org/10.3389/fbioe.2021.620777.
Peng H, He L, Haritos VS. Enhanced production of high-value cyclopropane fatty acid in yeast engineered for increased lipid synthesis and accumulation. Biotechnol J. 2019;14(4):1800487. https://doi.org/10.1002/biot.201800487.
Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. Genetic engineering of algae for enhanced biofuel production. Eukaryot Cell. 2010;9(4):486–501.
Rakicka M, Lazar Z, Dulermo T, Fickers P, Nicaud JM. Lipid production by the oleaginous yeast Yarrowia lipolytica using industrial by-products under different culture conditions. Biotechnol Biofuels. 2015;8:1–10.
Ramamurthy PC, Singh S, Kapoor D, Parihar P, Samuel J, Prasad R, Singh J. Microbial biotechnological approaches: renewable bioprocessing for the future energy systems. Microb Cell Fact. 2021;20(1):1–11.
Ratledge C, Wynn JP. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Advan Appl Microbiol. 2002;51:1–52.
Ratledge C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie. 2004;86(11):807–15.
Ratledge C. The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: a reappraisal and unsolved problems. Biotechnol Lett. 2014;36:1557–68.
Rengel R, Smith RT, Haslam RP, Sayanova O, Vila M, Leon R. Overexpression of acetyl- CoA synthetase (ACS) enhances the biosynthesis of neutral lipids and starch in the green microalga Chlamydomonas reinhardtii. Algal Res. 2018;31:183–93.
Rossi M, Amaretti A, Raimondi S, Leonardi A. Getting lipids for biodiesel production from oleaginous fungi. In: Stoytcheva M, Montero G, editors. Biodiesel-feedstocks and processing technologies. InTech; 2011. https://doi.org/10.5772/25864.
Rubin-Pitel SB, Cho CM, Chen W, Zhao H. Directed evolution tools in bioproduct and bioprocess development. In: Yang S, editor. Bioprocessing for value-added products from renewable resources. Elsevier; 2007. p. 49–72.
Saha R, Mukhopadhyay M. Prospect of metabolic engineering in enhanced microbial lipid production. Biomass Convers Biorefin. 2021. https://doi.org/10.1007/s13399-021-02114-4.
Saisriyoot M, Thanapimmetha A, Suwaleerat T, Chisti Y, Srinophakun P. Biomass and lipid production by Rhodococcus opacus PD630 in molasses-based media with and without osmotic-stress. J Biotechnol. 2019;297:1–8.
Santala S, Efimova E, Koskinen P, Karp MT, Santala V. Rewiring the wax ester production pathway of Acinetobacter baylyi ADP1. ACS Synth Biol. 2014;3(3):145–51.
Santek MI, Beluhan S, Santek B. Production of microbial lipids from lignocellulosic biomass. In: Rao MN, Soneji J, editors. Advances biofuels bioenergy. IntechOpen; 2018. p. 137–64.
Schaechter M. Encyclopedia of microbiology. Academic Press; 2009.
Shabbir Hussain M, Wheeldon I, Blenner MA. A strong hybrid fatty acid inducible transcriptional sensor built from Yarrowia lipolytica upstream activating and regulatory sequences. Biotechnol J. 2017;12(10):1700248.
Shah AM, Mohamed H, Fazili ABA, Yang W, Song Y. Investigating the effect of alcohol dehydrogenase gene knockout on lipid accumulation in Mucor circinelloides WJ11. J Fungi. 2022;8(9):917. https://doi.org/10.3390/jof8090917.
Shaigani P, Awad D, Redai V, Fuchs M, Haack M, Mehlmer N, Brueck T. Oleaginous yeasts-substrate preference and lipid productivity: a view on the performance of microbial lipid producers. Microbial Cell Fact. 2021;20(1):1–18.
Sharma KK, Schuhmann H, Schenk PM. High lipid induction in microalgae for biodiesel production. Energies. 2012;5(5):1532–53.
Singh Saharan B, Grewal A, Kumar P. Biotechnological production of polyhydroxyalkanoates: a review on trends and latest developments. Chinese J Biol. 2014. https://doi.org/10.1155/2014/802984.
Sohlenkamp C, Geiger O. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol Rev. 2016;40(1):133–59.
Soltani M, Metzger P, Largeau C. Fatty acid and hydroxy acid adaptation in three gram- negative hydrocarbon-degrading bacteria in relation to carbon source. Lipids. 2005;40(12):1263–72.
Somashekar D, Venkateshwaran G, Sambaiah K, Lokesh BR. Effect of culture conditions on lipid and gamma-linolenic acid production by mucoraceous fungi. Proc Biochem. 2003;38(12):1719–24.
Subramaniam R, Dufreche S, Zappi M, Bajpai R. Microbial lipids from renewable resources: production and characterization. J Ind Microbiol Biotechnol. 2010;37(12):1271–87.
Sun X, Li P, Liu X, Wang X, Liu Y, Turaib A, Cheng Z. Strategies for enhanced lipid production of Desmodesmus sp. mutated by atmospheric and room temperature plasma with a new efficient screening method. J Clean Product. 2020. https://doi.org/10.1016/j.jclepro.2019.119509.
Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metabol Eng. 2013;15:1–9.
Tanimura A, Takashima M, Sugita T, Endoh R, Kikukawa M, Yamaguchi S, Shima J. Cryptococcus terricola is a promising oleaginous yeast for biodiesel production from starch through consolidated bioprocessing. Sci Rep. 2014;4(1):1–6.
Tapia E, Anschau A, Coradini AL, Franco TT, Deckmann AC. Optimization of lipid production by the oleaginous yeast Lipomyces starkeyi by random mutagenesis coupled to cerulenin screening. AMB Express. 2012;2(1):1–8.
Trehan A, Kiełbus M, Czapinski J, Stepulak A, Huhtaniemi I, Rivero-Müller A. REPLACR-mutagenesis, a one-step method for site-directed mutagenesis by recombineering. Sci Rep. 2016;6(1):19121
Trentacoste EM, Shrestha RP, Smith SR, Glé C, Hartmann AC, Hildebrand M, Gerwick WH. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc Natl Acad Sci. 2013;110(49):19748–53.
Tsakraklides V, Kamineni A, Consiglio AL, MacEwen K, Friedlander J, Blitzblau HG, Brevnova EE. High-oleate yeast oil without polyunsaturated fatty acids. Biotechnol Biofuels. 2018;11(1):1–11.
Voelker TA, Davies HM. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J Bacteriol. 1994;176(23):7320–7.
Wakeham SG, Pease TK, Benner R. Hydroxy fatty acids in marine dissolved organic matter as indicators of bacterial membrane material. Org Geochem. 2003;34(6):857–68.
Wältermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Steinbüchel A. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol. 2005;55(3):750–63.
Wang J, Peng J, Fan H, Xiu X, Xue L, Wang L, Wang R. Development of mazF-based markerless genome editing system and metabolic pathway engineering in Candida tropicalis for producing long-chain dicarboxylic acids. J Ind Microbiol Biotechnol. 2018;45(11):971–81.
Wang S, Wan W, Wang Z, Zhang H, Liu H, Arunakumara KKIU, Song X. A two-stage adaptive laboratory evolution strategy to enhance docosahexaenoic acid synthesis in oleaginous thraustochytrid. Front Nutr. 2021;8:795491. https://doi.org/10.3389/fnut.2021.795491.
Wang X, Dong HP, Wei W, Balamurugan S, Yang WD, Liu JS, Li HY. Dual expression of plastidial GPAT1 and LPAT1 regulates triacylglycerol production and the fatty acid profile in Phaeodactylum tricornutum. Biotechnol Biofuels. 2018;11(1):1–14.
Wang XW, Liang JR, Luo CS, Chen CP, Gao YH. Biomass, total lipid production, and fatty acid composition of the marine diatom Chaetoceros muelleri in response to different CO2 levels. Bioresour Technol. 2014;161:124–30.
Wei H, Wang W, Alper HS, Xu Q, Knoshaug EP, Van Wychen S, Zhan M. Ameliorating the metabolic burden of the co-expression of secreted fungal cellulases in a high lipid- accumulating Yarrowia lipolytica strain by medium C/N ratio and a chemical chaperone. Front Microbiol. 2019. https://doi.org/10.3389/fmicb.2018.03276.
Wong YK, Ho YH, Ho KC, Leung HM, Chow KP, Yung KKL. Effect of different light sources on algal biomass and lipid production in internal leds-illuminated photobioreactor. J Mar Biol Aquacult. 2016;2(2):1–8.
Wu S, Hu C, Jin G, Zhao X, Zhao ZK. Phosphate-limitation mediated lipid production by Rhodosporidium toruloides. Bioresour Technol. 2010;101(15):6124–9.
Wynn JP, Hamid AA, Li Y, Ratledge C. Biochemical events leading to the diversion of carbon into storage lipids in the oleaginous fungi Mucor circinelloides and Mortierella alpina. Microbiology. 2001;147(10):2857–64.
Xin F, Dang W, Chang Y, Wang R, Yuan H, Xie Z, Song Y. Transcriptomic analysis revealed the differences in lipid accumulation between spores and mycelia of Mucor circinelloides WJ11 under solid–state fermentation. Fermentation. 2022;8(12):667.
Xin Y, Shen C, She Y, Chen H, Wang C, Wei L, Xu J. Biosynthesis of triacylglycerol molecules with a tailored PUFA profile in industrial microalgae. Mol Plant. 2019;12(4):474–88.
Xing W, Zhang R, Shao Q, Meng C, Wang X, Wei Z, Gao Z. Effects of laser mutagenesis on microalgae production and lipid accumulation in two economically important fresh Chlorella strains under heterotrophic conditions. Agronomy. 2021;11(5):961. https://doi.org/10.3390/agronomy11050961.
Xu W, Jiang X, Huang L. RNA interference technology. Comprehen Biotechnol. 2019. https://doi.org/10.1016/B978-0-444-64046-8.00282-2.
Xue J, Balamurugan S, Li DW, Liu YH, Zeng H, Wang L, Li HY. Glucose-6-phosphate dehydrogenase as a target for highly efficient fatty acid biosynthesis in microalgae by enhancing NADPH supply. Metabol Eng. 2017;41:212–21.
Yan FX, Dong GR, Qiang S, Niu YJ, Hu CY, Meng YH. Overexpression of o 12, o 15- desaturases for enhanced lipids synthesis in Yarrowia lipolytica. Front Microbiol. 2020;11:289. https://doi.org/10.3389/fmicb.2020.00289.
Yan J, Kuang Y, Gui X, Han X, Yan Y. Engineering a malic enzyme to enhance lipid accumulation in Chlorella protothecoides and direct production of biodiesel from the microalgal biomass. Biomass Bioenergy. 2019;122:298–304.
Yang Y, Hu B. Investigation on the cultivation conditions of a newly isolated fusarium fungal strain for enhanced lipid production. Appl Biochem Biotechnol. 2019;187:1220–37.
Yang J, Astatkie T, He QS. A comparative study on the effect of unsaturation degree of camelina and canola oils on the optimization of bio-diesel production. Energy Rep. 2016;2:211–7.
Yang L, Chen J, Qin S, Zeng M, Jiang Y, Hu L, Wang J. Growth and lipid accumulation by different nutrients in the microalga Chlamydomonas reinhardtii. Biotechnol Biofuels. 2018;11(1):1–12.
Yao Y, Lu Y, Peng KT, Huang T, Niu YF, Xie WH, Li HY. Glycerol and neutral lipid production in the oleaginous marine diatom Phaeodactylum tricornutum promoted by overexpression of glycerol-3-phosphate dehydrogenase. Biotechnol Biofuels. 2014;7(1):1–9.
Ying K, Zimmerman WB, Gilmour DJ. Effects of CO and pH on growth of the microalga Dunaliella salina. J Microb Biochem Technol. 2014;6(3):167–73.
Yoshida K, Hashimoto M, Hori R, Adachi T, Okuyama H, Orikasa Y, Morita N. Bacterial long-chain polyunsaturated fatty acids: their biosynthetic genes, functions, and practical use. Mar Drugs. 2016;14(5):94.
Yu T, Zhou YJ, Huang M, Liu Q, Pereira R, David F, Nielsen J. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell. 2018;174(6):1549–58.
Zavala-Moreno A, Arreguin-Espinosa R, Pardo JP, Romero-Aguilar L, Guerra-Sánchez G. Nitrogen source affects glycolipid production and lipid accumulation in the phytopathogen fungus Ustilago maydis. Advan Microbiol. 2014;4(13):934.
Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30(4):354–9.
Zhang L, Xiu X, Wang Z, Jiang Y, Fan H, Su J, Wang J. Increasing long-chain dicarboxylic acid production in Candida tropicalis by engineering fatty transporters. Mol Biotechnol. 2021;63(6):544–55.
Zhang Y, Adams IP, Ratledge C. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology. 2007;153(7):2013–25.
Zhang Y, Adams IP, Ratledge C. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology. 2007;153(7):2013–25.
Zhang Q, Li Y, Xia L. An oleaginous endophyte Bacillus subtilis HB1310 isolated from thin-shelled walnut and its utilization of cotton stalk hydrolysate for lipid production. Biotechnol Biofuel. 2014;7(1):1–13.
Zhao X, Peng F, Du W, Liu C, Liu D. Effects of some inhibitors on the growth and lipid accumulation of oleaginous yeast Rhodosporidium toruloides and preparation of biodiesel by enzymatic transesterification of the lipid. Bioproc Biosyst Eng. 2012;35(6):993–1004.
Zhu LD, Li ZH, Hiltunen E. Strategies for lipid production improvement in microalgae as a biodiesel feedstock. BioMed Res Int. 2016. https://doi.org/10.1155/2016/8792548.
Zhu S, Bonito G, Chen Y, Du Z. Oleaginous fungi in biorefineries. Encyclopedia Mycol. 2021;2:577–89.
Zhu Z, Hu Y, Teixeira PG, Pereira R, Chen Y, Siewers V, Nielsen J. Multidimensional engineering of Saccharomyces cerevisiae for efficient synthesis of medium-chain fatty acids. Nat Catal. 2020;3(1):64–74.
Acknowledgements
The authors are thankful to the SVMCM Scholarship funded by the Ministry of Higher Education, Govt. of West Bengal for the financial contribution.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
SM: conceptualization, data curation, writing—original draft. SKH: conceptualization, supervision, writing—review and editing. KCM: conceptualization, resources, funding acquisition, supervision, writing—review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mondal, S., Halder, S.K. & Mondal, K.C. State-of-art engineering approaches for ameliorated production of microbial lipid. Syst Microbiol and Biomanuf 4, 20–38 (2024). https://doi.org/10.1007/s43393-023-00195-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-023-00195-y