Abstract
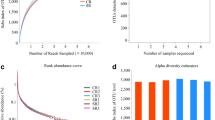
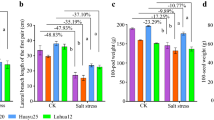
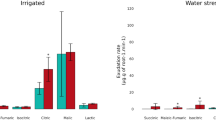
Soil salinization is a major constraint to crop cultivation. To improve the use-efficiency of salinized soil, sorghum and peanut are generally considered to be suitable crops for cultivation in these marginal soils. Herein, an artificially simulated salt-stress pot experiment was performed to investigate the effects of salt stress on microbial composition and metabolic profile in rhizosphere soil of sorghum and peanut, respectively. As a consequence, salt stress increased specific metabolites and recruitment of key microbial taxa in sorghum and peanut rhizosphere soils. Sorghum recruits key microbial taxa (including Nocardioides, Massilia, Gemmatirosa, and Pseudolabrys); it also regulates microbial genes encoding enzymes relevant to S_TMSMW0211 (n-Hexadecanoic, Acid) to participate in the fatty acid elongation pathway for salt stress response. The key microbial taxa (including Sphingomonas, and Gemmatimonadetes bacterium) recruited by peanut primarily mediate starch and sucrose metabolism pathway by enabling carbohydrate-related enzymes to cope with salt stress. Generally, the recruited microbial taxa and specific metabolites produced by sorghum and peanut under salt stress are associated with energy metabolism. This indicates that microbes potentially help their host cope with salt stress by mediating energy supply. This study revealed different strategies for salt stress response between the two crops at the microbiological level. The findings may help in identifying beneficial microbes that help plants cope with salt stress. Our findings can also provide novel perspectives for optimizing saline soil cultivation from the microbial management perspective.







Similar content being viewed by others
Data Availability
Data has been included in the manuscript.
References
Badri DV, Quintana N, El Kassis EG, Kim HK, Choi YH, Sugiyama A et al (2009) An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol 151:2006–2017. https://doi.org/10.1104/pp.109.147462
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. https://doi.org/10.1146/annurev.arplant.57.032905.105159
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. https://doi.org/10.1038/nmeth.3176
Calone R, Sanoubar R, Lambertini C, Speranza M, Antisari LV, Vianello G, Barbanti L (2020) Salt tolerance and na allocation in Sorghum bicolor under variable soil and water salinity. Plants 9:561. https://doi.org/10.3390/plants9050561
Cui F, Sui N, Duan G, Liu Y, Han Y, Liu S, Wan S, Li G (2018) Identification of metabolites and transcripts involved in salt stress and recovery in peanut. Front Plant Sci 9:217–264. https://doi.org/10.3389/fpls.2018.00217
Dodd IC, Pérez-Alfocea F (2012) Microbial amelioration of crop salinity stress. J Exp Bot 63:3415–3428. https://doi.org/10.1093/jxb/ers033
Etesami H, Maheshwari DK (2018) Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotox Environ Safe 156:225–246. https://doi.org/10.1016/j.ecoenv.2018.03.013
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Gou M, Qu Y, Yang H, Zhou J, Li A, Guan X, Ai F (2008) Sphingomonas sp.: a novel microbial resource for biodegradation of aromatic compounds. Chin J Appl Environ Biol 14:276–282. https://doi.org/10.3321/j.issn:1006-687X.2008.02.027(inChinese)
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Huang R (2018) Research progress on plant tolerance to soil salinity and alkalinity in sorghum. J Integr Agric 17:739–746. https://doi.org/10.1016/S2095-3119(17)61728-3
Huang L, Wu DZ, Zhang GP (2020) Advances in studies on ion transporters involved in salt tolerance and breeding crop cultivars with high salt tolerance. J Zhejiang Univ -SCI B 21:426–441. https://doi.org/10.1631/jzus.B1900510
Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC (2011) Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560. https://doi.org/10.1101/gr.120618.111
Ji C, Mao X, Hao J, Wang X, Xue J, Cui H, Li R (2018) Analysis of bZIP transcription factor family and their expressions under salt stress in Chlamydomonas reinhardtii. Int J Mol Sci 19:2800. https://doi.org/10.3390/ijms19092800
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Keswani C, Prakash O, Bharti N, Vílchez JI, Sansinenea E, Lally RD et al (2019) Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci Total Environ 690:841–852. https://doi.org/10.1016/j.scitotenv.2019.07.046
Kırtel O, Versluys M, Van den Ende W, Toksoy Öner E (2018) Fructans of the saline world. Biotechnol Adv 36(5):1524–1539. https://doi.org/10.1016/j.biotechadv.2018.06.009
Li D, Liu CM, Luo R, Sadakane K, Lam TW (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. https://doi.org/10.1093/bioinformatics/btv033
Li H, La S, Zhang X, Gao L, Tian Y (2021) Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J 15:2865–2882. https://doi.org/10.1038/s41396-021-00974-2
Lian T, Huang Y, Xie X, Huo X, Shahid MQ, Tian L, Lan T, Jin J (2020) Rice SST variation shapes the rhizosphere bacterial community, conferring tolerance to salt stress through regulating soil metabolites. mSystems 5. https://doi.org/10.1128/mSystems.00721-20
Liu Q, Tang J, Wang W, Zhang Y, Yuan H, Huang S (2018) Transcriptome analysis reveals complex response of the medicinal/ornamental halophyte Iris halophila Pall. to high environmental salinity. Ecotox Environ Safe 165:250–260. https://doi.org/10.1016/j.ecoenv.2018.09.003
Liu Y, Xun W, Chen L, Xu Z, Zhang N, Feng H, Zhang Q, Zhang R (2022) Rhizosphere microbes enhance plant salt tolerance: Toward crop production in saline soil. Comp Struct Biotechnol J 20:6543–6551. https://doi.org/10.1016/j.csbj.2022.11.046
Luo Q, Sun L, Hu X (2015) Metabonomics study on root exudates of cadmium hyperaccumulator Sedum alfredii. Chin J Anal Chem 43:7–12. https://doi.org/10.1016/S1872-2040(15)60795-2(inChinese)
Mertens J, Pollier J, Vanden Bossche R, Lopez-Vidriero I, Franco-Zorrilla JM, Goossens A (2016) The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol 170:194–210. https://doi.org/10.1104/pp.15.01645
Neal AL, Ahmad S, Gordon-Weeks R, Ton J (2012) Benzoxazinoids in root exudates of maize attract pseudomonas putida to the rhizosphere. PLoS ONE 7:e35498. https://doi.org/10.1371/journal.pone.0035498
Oburger E, Staudinger C, Spiridon A, Benyr V, Aleksza D, Wenzel W, Santangeli M (2022) A quick and simple spectrophotometric method to determine total carbon concentrations in root exudate samples of grass species. Plant Soil 478:273–281. https://doi.org/10.1007/s11104-022-05519-w
O'Neal L, Vo L, Alexandre G (2020) Specific root exudate compounds sensed by dedicated chemoreceptors shape azospirillum brasilense chemotaxis in the rhizosphere. Appl Environ Microbiol 86. https://doi.org/10.1128/aem.01026-20
Pisinaras V, Tsihrintzis VA, Petalas C, Ouzounis K (2010) Soil salinization in the agricultural lands of Rhodope District, northeastern Greece. Environ Monit Assess 166:79–94. https://doi.org/10.1007/s10661-009-0986-6
Radomiljac JD, Whelan J, Van der Merwe M (2013) Coordinating metabolite changes with our perception of plant abiotic stress responses: emerging views revealed by integrative—omic analyses. Metabolites 3:761–786. https://doi.org/10.3390/metabo3030761
Robles P, Quesada V (2019) Transcriptional and post-transcriptional regulation of organellar gene expression (OGE) and its roles in plant salt tolerance. Int J Mol Sci 20:1056. https://doi.org/10.3390/ijms20051056
Rolfe SA, Griffiths J, Ton J (2019) Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr Opin Microbiol 49:73–82. https://doi.org/10.1016/j.mib.2019.10.003
Rudrappa T, Czymmek KJ, Paré PW, Bais HP (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148:1547–1556. https://doi.org/10.1104/pp.108.127613
Shao F, Zhang L, Wilson IW, Qiu D (2018) Transcriptomic analysis of betula halophila in response to salt stress. Int J Mol Sci 19. https://doi.org/10.3390/ijms19113412
Shi W, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ (2015) The 40-residue insertion in Vibrio cholerae FadR facilitates binding of an additional fatty acyl-CoA ligand. Nat Commun 6:6032. https://doi.org/10.1038/ncomms7032
Shi X, Zhou Y, Guo P, Ren J, Zhang H, Dong Q, Jiang C, Zhong C, Zhang Z, Wan S, Zhao X, Yu H (2023a) Peanut/sorghum intercropping drives specific variation in peanut rhizosphere soil properties and microbiomes under salt stress. Land Degrad Dev 34:736–750. https://doi.org/10.1002/ldr.4490
Shi X, Zhou Y, Zhao X, Guo P, Ren J, Zhang H, Dong Q, Zhang Z, Yu H, Wan S (2023b) Soil metagenome and metabolome of peanut intercropped with sorghum reveal a prominent role of carbohydrate metabolism in salt-stress response. Environ Exp Bot 209:105274. https://doi.org/10.1016/j.envexpbot.2023.105274
Talaat NB, Shawky BT (2022) Synergistic effects of salicylic acid and melatonin on modulating ion homeostasis in salt-stressed wheat (Triticum aestivum L.) plants by enhancing root H+-pump activity. Plants 11:416. https://doi.org/10.3390/plants11030416
Upchurch RG (2008) Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol Lett 30:967–977. https://doi.org/10.1007/s10529-008-9639-z
Wang H, Sun T, Song W, Guo X, Cao P, Xu X, Shen Y, Zhao J (2020) Taxonomic characterization and secondary metabolite analysis of NEAU-wh3-1: An Embleya strain with antitumor and antibacterial activity. Microorganisms 8:441. https://doi.org/10.3390/microorganisms8030441
Williams A, Langridge H, Straathof AL, Fox G, Muhammadali H, Hollywood KA, Xu Y, Goodacre R, de Vries FT (2021) Comparing root exudate collection techniques: An improved hybrid method. Soil Biol Biochem 161:108391. https://doi.org/10.1016/j.soilbio.2021.108391
Xu L, Naylor D, Dong Z, Simmons T, Pierroz G, Hixson KK et al (2018) Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc Natl Acad Sci USA 115(18):E4284–E4293. https://doi.org/10.1073/pnas.1717308115
Yang ED, Cui DX, Wang WY (2019) Research progress on the genus Massilia. Microbiol China 46:1537–1548. https://doi.org/10.13344/j.microbiol.china.180573 (in Chinese)
Zhao X, Huang J, Lu J, Sun Y (2019) Study on the influence of soil microbial community on the long-term heavy metal pollution of different land use types and depth layers in mine. Ecotox Environ Safe 170:218–226. https://doi.org/10.1016/j.ecoenv.2018.11.136
Zhu W, Lomsadze A, Borodovsky M (2010) Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:132–146. https://doi.org/10.1093/nar/gkq275
Acknowledgements
This work were financially supported by the National Natural Science Foundation Youth Project of China (No. 32301948) and the China Agricultural Research System (No. CARS-06, CARS-13).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing any commercial or financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, X., Guo, P., Chen, Y. et al. Integrated Analysis of Soil Metagenome and Soil Metabolome Reveals the Differential Responses of Sorghum and Peanut Rhizosphere Microbes to Salt Stress. J Soil Sci Plant Nutr (2024). https://doi.org/10.1007/s42729-024-01721-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42729-024-01721-0