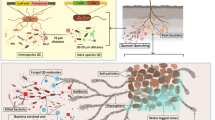
Abstract
Purpose of Review
Microbial interaction determines the performance, stability, and resilience of the microbial communities. Understanding microbial interactions benefits the development of environmental biotechnology. The purpose of this review is to summarize the recent findings of microbial interactions in pollution control ecosystems from aspects of the substrate degradation, energy utilization, electron transfer, and information exchange.
Recent Findings
Cross-feeding of substrates such as vitamins was found to be necessary for the degradation of some trace organic contaminants. Under different conditions, microorganisms can mediate various energy-utilization pathways (e.g., co-metabolism) to grow. Electroactive bacteria and cable bacteria can mediate extracellular electron transfer via conductive pili, c-type cytochrome, or filamentous structure. Quorum sensing plays an important role in the microbial aggregation and functional microbe enrichments. With all these knowledges, it will potentially benefit the development of disruptive environmental biotechnologies.
Summary
This review summarized recent findings of microbial interactions, many of which have huge potentials to advance environmental biotechnologies. Multi-omics methods should be further applied for comprehensively confirming known and unknown microbial processes. The co-occurrence network should be applied to unravel the interlinks among substrate degradation, energy utilization, electron transfer, and information exchange. The proper regulation of microbial interactions in practical application should be further addressed.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Bernstein HC, Carlson RP. Microbial consortia engineering for cellular factories: in vitro to in silico systems. Comput Struct Biotec. 2012;3(4):e201210017. https://doi.org/10.5936/csbj.201210017.
Biebl H, Pfenning N. Growth yields of green sulfur bacteria in mixed cultures with sulfur and sulfate reducing bacteria. Arch Microbiol. 1978;117:9–16. https://doi.org/10.1007/BF00689344.
Stams A, Plugge C. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol. 2009;7:568–77. https://doi.org/10.1038/nrmicro2166.
Morris BEL, Henneberger R, Huber H, Moissl-Eichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37(3):384–406. https://doi.org/10.1111/1574-6976.12019.
Hillesland KL. Evolution on the bright side of life: microorganisms and the evolution of mutualism. Ann NY Acad Sci. 2018;1422(1):88–103. https://doi.org/10.1111/nyas.13515.
Embree M, Liu JK, Al-Bassam MM, Zengler K. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc Natl Acad Sci USA. 2015;112(50):15450–5. https://doi.org/10.1073/pnas.1506034112.
Mori M, Ponce-de-León M, Peretó J, Montero F. Metabolic complementation in bacterial communities: necessary conditions and optimality. Front Microbiol. 2016;7:1553. https://doi.org/10.3389/fmicb.2016.01553.
Walker CB, Redding-Johanson AM, Baidoo EE, Rajeev L, He Z, Hendrickson EL, et al. Functional responses of methanogenic archaea to syntrophic growth. ISME J. 2012;6:2045–55. https://doi.org/10.1038/ismej.2012.60.
Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA. 2014;111(20):E2149–56. https://doi.org/10.1073/pnas.1405641111.
Walker DJF, Adhikari RY, Holmes DE, Ward JE, Woodard TL, Nevin KP, et al. Electrically conductive pili from pilin genes of phylogenetically diverse microorganisms. ISME J. 2017;12(1):48–58. https://doi.org/10.1038/ismej.2017.141.
Marozava S, Mouttaki H, Müller H, Laban NA, Probst AJ, Meckenstock RU. Anaerobic degradation of 1-methylnaphthalene by a member of the Thermoanaerobacteraceae contained in an iron-reducing enrichment culture. Biodegradation. 2018;29(1):23–39. https://doi.org/10.1007/s10532-017-9811-z.
• Wiechmann A, Ciurus S, Oswald F, Seiler VN, Müller V. It does not always take two to tango: “Syntrophy” via hydrogen cycling in one bacterial cell. ISME J. 2020;14:1561–70. https://doi.org/10.1038/s41396-020-0627-1This article first describes a novel type of hydrogen cycling that connects an oxidative and reductive metabolic module in one bacterial cell, “intracellular syntrophy.”.
• Leu AO, Cai C, McIlroy SJ, Southam G, Orphan VJ, Yuan Z, et al. Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J. 2020;14:1030–41. https://doi.org/10.1038/s41396-020-0590-xThis article describes the coupling of anaerobic methane oxidation and manganese reduction, implying the potential role of Methanoperedenaceae in linking methane and manganese cycling in the environment.
Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature. 2015;526:587–90. https://doi.org/10.1038/nature15733.
Lackner S, Gilbert EM, Vlaeminck SE, Joss A, Horn H, van Loosdrecht MCM. Full-scale partial nitritation/anammox experiences - an application survey. Water Res. 2014;55:292–303. https://doi.org/10.1016/j.watres.2014.02.032.
Zhou J, He Q, Hemme CL, Mukhopadhyay A, Hillesland K, Zhou A, et al. How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat Rev Microbiol. 2011;9(6):452–66. https://doi.org/10.1038/nrmicro2575.
Speth DR, in’t Zandt MH, Guerrero-Cruz S, Dutilh BE, Jetten MSM. Genomebased microbial ecology of anammox granules in a full-scale wastewater treatment system. Nat Commun. 2016;7:11172. https://doi.org/10.1038/ncomms11172.
Jung H, Baek G, Lee C. Magnetite-assisted in situ microbial oxidation of H2S to S0 during anaerobic digestion: a new potential for sulfide control. Chem Eng J. 2020;397:124982. https://doi.org/10.1016/j.cej.2020.124982.
Barret M, Delgadillo-Mirquez L, Trably E, Delgenès N, Braun F, Cea-Barcia G, et al. Anaerobic removal of trace organic contaminants in sewage sludge: 15 years of experience. Pedosphere. 2012;22(4):508–17. https://doi.org/10.1016/S1002-0160(12)60035-6.
Wang B, Liu W, Zhang Y, Wang A. Intermittent electro field regulated mutualistic interspecies electron transfer away from the electrodes for bioenergy recovery from wastewater. Water Res. 2020;185:116238. https://doi.org/10.1016/j.watres.2020.116238.
Elías-Arnanz M. Anaerobic bacteria need their vitamin B12 to digest estrogen. Proc Natl Acad Sci USA. 2020;117(4):1833–5. https://doi.org/10.1073/pnas.1921340117.
• Han P, Yu Y, Zhou L, Tian Z, Li Z, Hou L, et al. Specific micropollutant biotransformation pattern by the comammox bacterium Nitrospira inopinata. Environ Sci Technol. 2019;53(15):8695–705. https://doi.org/10.1021/acs.est.9b01037This article describes the micropollutant degradation potential of a comammox bacterium to understand the fate of micropollutants in nitrifying environments.
Koch H, van Kessel MA, Lücker S. Complete nitrification: insights into the ecophysiology of comammox Nitrospira. Appl Microbiol Biotechnol. 2019;103(1):177–89. https://doi.org/10.1007/s00253-018-9486-3.
Palomo A, Pedersen AG, Fowler SJ, Dechesne A, Sicheritz-Pontén T, Smets BF. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 2018;12(7):1779–93. https://doi.org/10.1038/s41396-018-0083-3.
Von Wintersdorff CJ, Penders J, Van Niekerk JM, Mills ND, Majumder S, Van Alphen LB, et al. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol. 2016;7:173. https://doi.org/10.3389/fmicb.2016.00173.
Criddle CS. The kinetics of cometabolism. Biotechnol Bioeng. 1993;41(11):1048–56. https://doi.org/10.1002/bit.260411107.
Sun Y, Guan Y, Wang D, Liang K, Wu G. Potential roles of acyl homoserine lactone based quorum sensing in sequencing batch nitrifying biofilm reactors with or without the addition of organic carbon. Bioresour Technol. 2018;259:136–45. https://doi.org/10.1016/j.biortech.2018.03.025.
•• Sandfeld T, Marzocchi U, Petro C, Schramm A, Risgaard-Petersen N. Electrogenic sulfide oxidation mediated by cable bacteria stimulates sulfate reduction in freshwater sediments. ISME J. 2020;14:1233–46. https://doi.org/10.1038/s41396-020-0607-5This article shows that cable bacteria stimulate sulfate reduction in freshwater sediment through promotion of sulfate availability, providing a mechanism for sulfate recycling.
Ghattas A-K, Fischer F, Wick A, Ternes TA. Anaerobic biodegradation of (emerging) organic contaminants in the aquatic environment. Water Res. 2017;116:268–95. https://doi.org/10.1016/j.watres.2017.02.001.
Lovley DR. Reach out and touch someone: potential impact of DIET (direct interspecies energy transfer) on anaerobic biogeochemistry, bioremediation, and bioenergy. Rev Environ Sci Biotechnol. 2011;10:101–5. https://doi.org/10.1007/s11157-011-9236-9.
Huang J, Wen Y, Ding N, Xu Y, Zhou Q. Fast start-up and stable performance coupled to sulfate reduction in the nitrobenzene bio-reduction system and its microbial community. Bioresour Technol. 2012;114:201–6. https://doi.org/10.1016/j.biortech.2012.03.050.
Cornelissen G, Sijm DTHM. An energy budget model for the biodegradation and cometabolism of organic substances. Chemosphere. 1996;33(5):817–30. https://doi.org/10.1016/0045-6535(96)00237-8.
Jewell KS, Falås P, Wick A, Joss A, Ternes TA. Transformation of diclofenac in hybrid biofilm–activated sludge processes. Water Res. 2016;105:559–67. https://doi.org/10.1016/j.watres.2016.08.002.
Rotaru AE, Shrestha PM, Liu F, Markovaite B, Chen S, Nevin KP, et al. Direct interspecies electron transfer between Geobacter metallireducens and Methanosarcina barkeri. Appl Environ Microbiol. 2014;80(15):4599–605. https://doi.org/10.1128/AEM.00895-14.
Rotaru AE, Shrestha PM, Liu F, Shrestha M, Shrestha D, Embree M, et al. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci. 2014;7(1):408–15. https://doi.org/10.1039/c3ee42189a.
• Walker DJF, Nevin KP, Holmes DE, Rotaru A-E, Ward JE, Zhu J, et al. Syntrophus conductive pili demonstrate that common hydrogen-donating syntrophs can have a direct electron transfer option. ISME J. 2020;14:837–46. https://doi.org/10.1038/s41396-019-0575-9This article reports that hydrogen-donating syntrophs Syntrophus aciditrophicus could produce e-pili and can grow via DIET.
McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature. 2015;526:531–5. https://doi.org/10.1038/nature15512.
Cruz Viggi C, Rossetti S, Fazi S, Paiano P, Majone M, Aulenta F. Magnetite particles triggering a faster and more robust syntrophic pathway of methanogenic propionate degradation. Environ Sci Technol. 2014;48:7536–43. https://doi.org/10.1021/es5016789.
Yin Q, Wu G. Advances in direct interspecies electron transfer and conductive materials: electron flux, organic degradation and microbial interaction. Biotechnol Adv. 2019;37:107443. https://doi.org/10.1016/j.biotechadv.2019.107443.
Liu Y, Gu M, Yin Q, Wu G. Inhibition mitigation and ecological mechanism of mesophilic methanogenesis triggered by supplement of ferroferric oxide in sulfate containing systems. Bioresour Technol. 2019;288:121546. https://doi.org/10.1016/j.biortech.2019.121546.
Yin Q, Gu M, Wu G. Inhibition mitigation of methanogenesis processes by conductive materials: a critical review. Bioresour Technol. 2020;317:123977. https://doi.org/10.1016/j.biortech.2020.123977.
Zhuang L, Ma J, Yu Z, Wang Y, Tang J. Magnetite accelerates syntrophic acetate oxidation in methanogenic systems with high ammonia concentrations. Microb Biotechnol. 2018;11(4):710–20. https://doi.org/10.1111/1751-7915.13286.
Liu Y, Gu M, Yin Q, Du J, Wu G. Thermodynamic analysis of direct interspecies electron transfer in syntrophic methanogenesis based on the optimized energy distribution. Bioresour Technol. 2020;297:122345. https://doi.org/10.1016/j.biortech.2019.122345.
Gu M, Yin Q, Liu Y, Du J, Wu G. New insights into the effect of direct interspecies electron transfer on syntrophic methanogenesis through thermodynamic analysis. Bioresour Technol Rep. 2019;7:100225. https://doi.org/10.1016/j.biteb.2019.100225.
Pirbadian S, Chavez MS, El-Naggar MY. Spatiotemporal mapping of bacterial membrane potential responses to extracellular electron transfer. Proc Natl Acad Sci USA. 2020;117(33):20171–9. https://doi.org/10.1073/pnas.2000802117.
Martineza CM, Alvarezb LH. Application of redox mediators in bioelectrochemical systems. Biotechnol Adv. 2018;36:1412–23. https://doi.org/10.1016/j.biotechadv.2018.05.005.
Gurumurthy DM, Bharagava RN, Kumar A, Singh B, Ashfaq M, Saratale GD, et al. EPS bound flavins driven mediated electron transfer in thermophilic Geobacillus sp. Microbiol Res. 2019;229:126324. https://doi.org/10.1016/j.micres.2019.126324.
Dubé CD, Guiot SR. Ethanol-to-methane activity of Geobacter-deprived anaerobic granules enhanced by conductive microparticles. Process Biochem. 2017;63:42–8. https://doi.org/10.1016/j.procbio.2017.07.032.
• Saunders SH, Tse ECM, Yates MD, Otero FJ, Trammell SA, Stemp EDA, et al. Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell. 2020;182(4):919–32. https://doi.org/10.1016/j.cell.2020.07.006This article describes phenazines mediate efficient extracellular electron transfer through interactions with extracellular DNA in Pseudomonas aeruginosa biofilms.
Xu S, Jangir Y, El-Naggar MY. Disentangling the roles of free and cytochrome-bound flavins in extracellular electron transport from Shewanella oneidensis MR-1. Electrochim Acta. 2016;198:49–55. https://doi.org/10.1016/j.electacta.2016.03.074.
Edwards MJ, White GF, Norman M, Tome-Fernandez A, Ainsworth E, Shi L, et al. Redox linked flavin sites in extracellular decaheme proteins involved in microbe-mineral electron transfer. Sci Rep. 2015;5:11677. https://doi.org/10.1038/srep11677.
Okamoto A, Hashimoto K, Nealson KH, Nakamura R. Rate enhancement of bacterial extracellular electron transport involves bound flavin semiquinones. Proc Natl Acad Sci USA. 2013;110:7856–61. https://doi.org/10.1073/pnas.1220823110.
Zakaria BS, Dhar BR. Changes in syntrophic microbial communities, EPS matrix, and gene-expression patterns in biofilm anode in response to silver nanoparticles exposure. Sci Total Environ. 2020;734:139395. https://doi.org/10.1016/j.scitotenv.2020.139395.
Zhang Z, Qu Y, Li S, Feng K, Cai W, Yin H, et al. Florfenicol restructured the microbial interaction network for wastewater treatment by microbial electrolysis cells. Environ Res. 2020;183:109145. https://doi.org/10.1016/j.envres.2020.109145.
Cai W, Liu W, Zhang Z, Feng K, Ren G, Pu C. Electro-driven methanogenic microbial community diversity and variability in the electron abundant niche. Sci Total Environ. 2019;661:178–86. https://doi.org/10.1016/j.scitotenv.2019.01.131.
Wang PH, Chen YL, Wei STS, Wu K, Lee TH, Wu TY, et al. Retroconversion of estrogens into androgens by bacteria via a cobalamin-mediated methylation. Proc Natl Acad Sci USA. 2020;117(3):1395–403. https://doi.org/10.1073/pnas.1914380117.
Hasany M, Mardanpour MM, Yaghmaei S. Biocatalysts in microbial electrolysis cells: a review. Int J Hydrog Energy. 2016;41(3):1477–93. https://doi.org/10.1016/j.ijhydene.2015.10.097.
Chabert N, Ali OA, Achouak W. All ecosystems potentially host electrogenic bacteria. Bioelectrochemistry. 2015;106:88–96. https://doi.org/10.1016/j.bioelechem.2015.07.004.
•• Meysman FJR, Cornelissen R, Trashin S, Bonné R, Martinez SH, van der Veen J, et al. A highly conductive fibre network enables centimetre-scale electron transport in multicellular cable bacteria. Nat Commun. 2019;10:4120. https://doi.org/10.1038/s41467-019-12115-7This article describes the cable bacteria can conduct electrons over centimetre distances via highly conductive fibres embedded in the cell envelope and the charge transfer is electronic rather than ionic.
Marzocchi U, Trojan D, Larsen S, Meyer RJ, Schramm A, Nielsen LP, et al. Electric coupling between distant nitrate reduction and sulfide oxidation in marine sediment. ISME J. 2014;8:1682–90. https://doi.org/10.1038/ismej.2014.19.
Geerlings NMJ, Karman C, Trashin S, As KS, Kienhuis MVM, Hidalgo-Martinez S, et al. Division of labor and growth during electrical cooperation in multicellular cable bacteria. Proc Natl Acad Sci USA. 2020;117(10):5478–85. https://doi.org/10.1073/pnas.1916244117.
Müller M, Marozava S, Probst AJ, Meckenstock RU. Groundwater cable bacteria conserve energy by sulfur disproportionation. ISME J. 2020;14:623–34. https://doi.org/10.1038/s41396-019-0554-1.
Scholz VV, Meckenstock RU, Nielsen LP, Risgaard-Petersen N. Cable bacteria reduce methane emissions from rice-vegetated soils. Nat Commun. 2020;11:1878. https://doi.org/10.1038/s41467-020-15812-w.
Marzocchi U, Palma E, Rossetti S, Aulenta F, Scoma A. Parallel artificial and biological electric circuits power petroleum decontamination: the case of snorkel and cable bacteria. Water Res. 2020;173:115520. https://doi.org/10.1016/j.watres.2020.115520.
Ye Q, Zhang Z, Huang Y, Fang T, Cui Q, He C, et al. Enhancing electron transfer by magnetite during phenanthrene anaerobic methanogenic degradation. Int Biodeterior Biodegrad. 2018;129:109–16. https://doi.org/10.1016/j.ibiod.2018.01.012.
Yu L, Yuan Y, Tang J, Wang Y, Zhou S. Biochar as an electron shuttle for reductive dechlorination of pentachlorophenol by Geobacter sulfurreducens. Sci Rep. 2015;5:16221. https://doi.org/10.1038/srep16221.
Cruz-Zavala AS, Pat-Espadas AM, Rangel-Mendez JR, Chazaro-Ruiz LF, Ascacio-Valdes JA, Aguilar CN, et al. Immobilization of metal–humic acid complexes in anaerobic granular sludge for their application as solid-phase redox mediators in the biotransformation of iopromide in UASB reactors. Bioresour Technol. 2016;207:39–45. https://doi.org/10.1016/j.biortech.2016.01.125.
Liu C, Xu X, Fan J. Accelerated anaerobic dechlorination of DDT in slurry with Hydragric Acrisols using citric acid and anthraquinone-2,6-disulfonate (AQDS). J Environ Sci. 2015;38:87–94. https://doi.org/10.1016/j.jes.2015.05.005.
Lefevre E, Redfern L, Cooper EM, Stapleton HM, Gunsch CK. Acetate promotes microbial reductive debromination of tetrabromobisphenol A during the startup phase of anaerobic wastewater sludge bioreactors. Sci Total Environ. 2019;656:959–68. https://doi.org/10.1016/j.scitotenv.2018.11.403.
Ma H, Wang X, Zhang Y, Hu H, Ren H, Geng J, et al. The diversity, distribution and function of N-acyl-homoserine lactone (AHL) in industrial anaerobic granular sludge. Bioresour Technol. 2018;247:116–24. https://doi.org/10.1016/j.biortech.2017.09.043.
Wei W, Zhang Y, Komorek R, Plymale A, Yu R, Wang B, et al. Characterization of syntrophic Geobacter communities using ToF-SIMS. Biointerphases. 2017;12(5):05G601. https://doi.org/10.1116/1.4986832.
Zhang G, Zhang F, Ding G, Li J, Guo X, Zhu J, et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 2012;6(7):1336–44. https://doi.org/10.1038/ismej.2011.203.
Sun Y, Guan Y, Zeng D, He K, Wu G. Metagenomics-based interpretation of AHLs-mediated quorum sensing in Anammox biofilm reactors for low-strength wastewater treatment. Chem Eng J. 2018;344:42–52. https://doi.org/10.1016/j.cej.2018.03.047.
• Tommonaro G, Abbamondi GR, Iodice C, Tait K, De Rosa S. Diketopiperazines produced by the halophilic archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb Ecol. 2012;63:490–5. https://doi.org/10.1007/s00248-011-9980-yThis study first describes that archaea might be able to interact with AHL-producing bacteria within mixed communities.
Li L, Zheng M, Ma H, Gong S, Ai G, Liu X, et al. Significant performance enhancement of a UASB reactor by using acyl homoserine lactones to facilitate the long filaments of Methanosaeta harundinacea 6Ac. Appl Microbiol Biotechnol. 2015;99(15):6471–80. https://doi.org/10.1007/s00253-015-6478-4.
Lv L, Li W, Zheng Z, Li D, Zhang N. Exogenous acyl-homoserine lactones adjust community structures of bacteria and methanogens to ameliorate the performance of anaerobic granular sludge. J Hazard Mater. 2018;354:72–80. https://doi.org/10.1016/j.jhazmat.2018.04.075.
Bordeleau E, Purcell EB, Lafontaine DA, Fortier LC, Tamayo R, Burrus V. Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J Bacteriol. 2015;197(5):819–32. https://doi.org/10.1128/JB.02340-14.
Sun Y, He K, Yin Q, Echigo S, Wu G, Guan Y. Determination of quorum-sensing signal substances in water and solid phases of activated sludge systems using liquid chromatography–mass spectrometry. J Environ Sci. 2018;69:85–94. https://doi.org/10.1016/j.jes.2017.04.017.
Du Q, Mu Q, Wu G. Metagenomic and bioanalytical insights into quorum sensing of methanogens in anaerobic digestion systems with or without the addition of conductive filter. Sci Total Environ. 2021;763:144509. https://doi.org/10.1016/j.scitotenv.2020.144509.
Li FH, Tang Q, Fan YY, Li Y, Li J, Wu JH, et al. Developing a population-state decision system for intelligently reprogramming extracellular electron transfer in Shewanella oneidensis. Proc Natl Acad Sci USA. 2020;117(37):23001–10. https://doi.org/10.1073/pnas.2006534117.
Lovley DR. Happy together: microbial communities that hook up to swap electrons. ISME J. 2017;11:327–36. https://doi.org/10.1038/ismej.2016.136.
Farag IF, Biddle JF, Zhao R, Martino AJ, House CH. León-Zayas Metabolic potentials of archaeal lineages resolved from metagenomes of deep Costa Rica sediments. ISME J. 2020;14:1345–58. https://doi.org/10.1038/s41396-020-0615-5.
Bell E, Lamminmäki T, Lneberg J, Andersson AF, Qian C, Xiong W, et al. Active sulfur cycling in the terrestrial deep subsurface. ISME J. 2020;14:1260–72. https://doi.org/10.1038/s41396-020-0602-x.
van Tatenhove-Pel RJ, Rijavec T, Lapanje A, van Swam I, Zwering E, Hernandez-Valdes JA, et al. Microbial competition reduces metabolic interaction distances to the low μm-range. ISME J. 2020;15:1–14. https://doi.org/10.1038/s41396-020-00806-9.
Islam MA, Karim A, Mishra P, Dubowski JJ, Yousuf A, Sarmin S. Microbial synergistic interactions enhanced power generation in co-culture driven microbial fuel cell. Sci Total Environ. 2020;738:140138. https://doi.org/10.1016/j.scitotenv.2020.140138.
Acknowledgements
This research was supported by the Galway University Foundation, and the Shenzhen Science and Technology Innovation Committee (grant number JCYJ20170817161106801).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Water Pollution
Rights and permissions
About this article
Cite this article
Yin, Q., Feng, Z., Hu, Y. et al. Microbial Interactions in Pollution Control Ecosystems. Curr Pollution Rep 7, 104–114 (2021). https://doi.org/10.1007/s40726-021-00181-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40726-021-00181-9