Abstract
Background
Increasing environmental awareness is forcing waste creators to consider new options such as biosorption for the disposal of colored wastewaters. Due to prohibitive costs of commercially available activated carbon, low-cost biosorbents with high adsorption capacities have gained increasing attention. The present investigation deals with utilization of a low-cost, fungal biosorbent of Rhizopus arrhizus NCIM 997 and optimization of conditions for the removal of Reactive Orange 13 dye from an aqueous solution using sequential statistically designed experiments.
Results
Plackett–Burman design with six independent variables (pH, temperature, biosorbent dosage, dye concentration, contact time and speed of agitation) was used to identify the most important factors influencing dye biosorption. Path of steepest ascent and central composite design were used to move toward the vicinity of the optimum operating conditions and to determine the optimum levels of the variables, respectively. The maximum biosorption capacity (133.63 mg/g) was obtained under the following conditions: pH 2.0, dye concentration 114 mg/L, biosorbent dosage 0.8 g/L and speed of agitation 85 rpm. Validation experiments and application of artificial neural network showed excellent correlation between predicted and experimental values.
Conclusions
Response surface methodology using central composite design was employed at the specified combinations of four independent significant factors identified by Plackett–Burman design. The fitted model was used to arrive at the best operating conditions to achieve a maximum response. Sequential optimization was successfully used to increase biosorption by 49.04 %. The study indicated that the fungal biosorbent was an effective and economical alternative for the removal of Reactive Orange 13 dye.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
India’s dye industry produces various types of dyes, of which 50 % are reactive dyes [1, 2]. Approximately, 70 % of the synthetic dyes belong to the azo group and, unfortunately, this class of dyes is the most unfavorable from the ecological point of view, as the effluents are heavily colored, contain high concentrations of salt and exhibit high BOD/COD values. The release of these dyes is both esthetically unacceptable and presents an eco-toxic hazard [2–4]. Currently, textile effluents are treated by physico-chemical methods such as electrocoagulation, ion exchange, irradiation, ozonation and advanced oxidation, which are of limited use due to constraints such as costs, general applicability and production of solid wastes [5, 6]. Unfavorable conditions found in the textile dyeing effluents are known to inhibit the conventional biological wastewater treatment processes. Removal of color from the effluents is thus a major problem forcing industries to consider new options for the effluent treatment and disposal [3]. Among all the wastewater treatments, the adsorption process has been recognized to be an effective and economical procedure for the removal of dyes from industrial effluents [7]. Activated carbon has an excellent adsorption capacity for organic pollutants; its prohibitive cost and inability to regenerate limit its commercial application [8]. Hence, low-cost biosorbents with high adsorption capacities such as waste materials from large-scale industrial operations, natural materials derived from agriculture, microbial biosorbents such as Corynebacterium glutamicum, Escherichia coli, Pseudomonas luteola and Rhizopus arrhizus have gained attention [9, 10]. A major advantage of biosorption is that it can be used in situ and integrated with many systems in the most eco-friendly manner [11].
Some of the most important parameters that affect the efficiency of biosorbent are time, pH and temperature of the solution and concentration of the dye as well as that of the biosorbent. Hence, it is necessary to investigate the combined effect of these factors on biosorption efficiency. Designing of experiment and standardization of variables affecting the system are very critical in the optimization process. Generally, this optimization is carried out by using traditional one factor at a time method, which though simple consumes time and chemicals in large quantities. Moreover, this method neglects the interaction effects of process variables. Hence, in the present study, multi-variant experimental design based on statistical approach such as response surface methodology (RSM) and artificial neural network (ANN) approach was applied for process modeling. RSM is a collection of statistical techniques used to design experiments, build models, evaluate the effects of various factors and search the optimum conditions for the factors to achieve the desired responses. It has been widely used for wastewater treatment processes. Its application in the adsorption process can result in improved sorption, reduced process variability, reduced development time and overall costs [12, 13]. Recently, ANN methodologies are being used in many areas of science and engineering to solve environmental engineering problems such as chromium removal [14] and textile dye removal [15, 16]. ANNs are considered as promising tool because of their simplicity toward simulation, less time required for model development than the traditional mathematical models [17], accurate prediction ability with limited numbers of experiments and identification of optimal operating conditions for the plant operator [18].
Application of dead fungal biomass obtained from R. arrhizus to remove textile dyes from industrial waste water is attractive due to its cheap and constant supply from industrial fermentation processes, high removal rates, easy storage and regenerative potential [11]. Some researchers have demonstrated the high dye binding capacity of R. arrhizus which exceeds that of some commercial ion exchange resins or activated carbon [19–21]. Hence, the main objective of the present study was to apply RSM and ANN approach for techno-economical optimization of the removal of Reactive Orange 13 (RO 13) dye, a mono-azo reactive dye from an aqueous solution using the dead biomass of R. arrhizus NCIM 997 as a low-cost biosorbent. In this study, the optimum value of the parameters and interactions between factors that influence the removal of RO 13 dye were determined. RO 13 was chosen in this study due to its extensive use in Indian textile industry for dyeing and printing cotton, viscose, silk, wool and nylon fabric.
Methods
Preparation of the biosorbent
Rhizopus arrhizus NCIM 997, obtained from National Collection of Industrial Microorganisms (NCIM), National Chemical Laboratory, Pune, India, was used as a biosorbent. It was routinely maintained at 4 °C on potato dextrose agar (PDA) (g/L: potato infusion from 200 g potatoes, dextrose 20, yeast extracts 0.1, agar 20, pH 5.0). For experimental purposes, fungal mycelia were obtained by aseptically transferring mycelia from the PDA spread-plate cultures to 100 mL of potato dextrose broth (PDB) containing 0.25 % Tween 80 (to prevent sporulation) in 250 mL Erlenmeyer flasks. The flasks were incubated at 30 ± 1 °C under static conditions with intermittent shaking. The biomass harvested after 7 days was washed thoroughly with generous amounts of deionized distilled water and dried at 80 °C in an oven for 24 h. Care was taken to keep the particle size of the biomass uniform, by grinding into powder and sieving through a 150-mesh sieve. The biomass was stored in a desiccator until use for the biosorption experiments [22].
Preparation of the dye solution
RO 13 (C.I. 18270, CAS 12225-85-3, chemical formula: C24H15CIN7Na3O10S3, molecular weight: 762.04 g/mol) was supplied by Colourtex Dyes Pvt. Ltd. (Mumbai, India) and was used without further purification. A stock solution of RO 13 dye (1000 mg/L) was prepared by dissolving the required amount of dye powder in double deionized water. The stock solution was diluted with double deionized water as required to obtain the desired concentration ranging from 40 to 100 mg/L. The required pH was adjusted by 0.1 N HCl or 0.1 N NaOH using a pH meter (Labotronics, LT-10, India).
Dye biosorption experiments
Biosorption experiments were conducted at the specified combinations of physical parameters using sequential statistically designed experiments consisting of Plackett–Burman design (PBD), path of steepest ascent (PSA) and central composite design (CCD). The experiments were carried out in 250 mL Erlenmeyer flasks with working volume of 50 mL of the reaction mixture. The flasks were withdrawn from the rotating orbital shaker (Labtop Quality Lab Equipment, India) after shaking for the desired time of reaction. The residual dye concentration in the solution was determined after filtering the samples using Whatman No. 1 filter paper. The concentration of RO13 dye in the solution was determined from the calibration curve prepared by measuring the absorbance of different known concentrations of RO 13 dye at 489 nm using a UV–vis spectrophotometer (Shimadzu, Kyoto, Japan). A blank without biosorbent was run simultaneously as a control. The concentration of the dye on the fungal biomass at the corresponding equilibrium conditions was determined using a mass balance equation expressed as specific uptake capacity (SUC):
where q is the amount of dye adsorbed per unit weight of the biosorbent (mg/g); C 0 the initial concentration of the dye (mg/L); C e the concentration of dye in solution at equilibrium time (mg/L); V the solution volume (L); m the dosage of the biosorbent (g). All the experiments were carried out in duplicate and the mean values are presented.
Experimental design and data analysis
Screening of parameters using PBD
A 12-run PBD along with three runs at zero level (in duplicate) was used in the present study to screen the important variables that significantly influenced biosorption. Six variables, viz. pH, dye concentration, biosorbent dosage, temperature, speed of agitation and contact time, were chosen as the independent input variables and the efficiency of dye removal was used as a dependent response variable. The center point replicates were chosen to verify any change in the estimation procedure as a measure of precision property. Each variable was examined at two levels: −1 for the low level and +1 for the high level (Table 1) [23].
Independent variables, experimental range and levels for RO 13 dye removal are given in Table 2.
The effect of individual variable on biosorption was calculated by Eq. (1)
where E (X i ) is the effect of the tested variable (X i ) and M +i and M −i are responses (biosorption) of trials at which the variable is at its high or low level, respectively. N is the total number of trials. From the regression analysis, significant factors affecting biosorption were determined and the contribution of the factors toward the sorption of RO 13 dye was determined based on the t value (main effect). The sign of the effect indicates the level at which it is considered for further improvement.
Locating the region of optimum response by the PSA
The factors that were screened using the PBD were further optimized using the PSA to move toward the vicinity of the optimum results. To improve biosorption, concentrations of variables were increased or decreased using stepwise units according to the sign of the main effects. The zero level of PBD was identified as the base point of PSA and, for every point in the PSA, an experimental run was performed. The step along the path was determined by practical experience. Experiments were performed along the steepest ascent path until the response showed no further increase. This point would be near the optimal point and could be used as the center point of CCD [24].
Optimization by response surface methodology using CCD
The optimal levels of the significant factors and the interactions of these variables on biosorption were analyzed by using CCD [17]. A four-factor, five-level CCD with 30 runs was conducted in the optimum vicinity to locate the true optimum values of pH (X 1), dye concentration (X 2), biosorbent dosage (X 3) and speed of agitation (X 5) combining factorial points (−1, +1), axial points (−2, +2) and central point (0).
The factors were coded according to the following Eq. (2):
where X i is the coded independent factor, x i the real independent factor and x 0 the value of X i at the center point; Δx is the step change value. For statistical calculations, the variables X i are coded as x i.
A second-order polynomial response equation (Eq. 3) was proposed to correlate the dependent and independent variables:
where Y is the predicted response, β 0 is the intercept, xi and x j are the coded independent factors, β i is the linear coefficient, β ii is the quadratic coefficient and βij is the interaction coefficient.
The quality of polynomial equation was judged by the determination coefficient (R 2) and its statistical significance was checked by Fischer’s F test. Analysis of variance (ANOVA) was conducted to determine the significance of the model. The response surface plots of the model-predicted responses were utilized to assess the interactive relationships between the significant variables. To verify the results obtained from the statistical analysis of CCD, validation tests were performed in duplicate using the predicted optimized conditions against the basal conditions.
Data analysis
Minitab 16 (State College, PA, USA) and Design Expert version 6.0.8 (Stat-Ease Inc., Minneapolis, USA) software were used for designing experiments as well as for regression and graphical analysis of the experimental data obtained.
Results and discussion
Screening of parameters using PBD
Plackett–Burman analysis is a two-level fractional factorial design for screening and evaluation of critical variables which might significantly affect the end response, while eliminating the insignificant variables. It allows unbiased estimation of main effects with smallest possible variance of components [23, 25]. The data listed in Table 2 indicate a wide variation in the specific uptake capacity (SUC) of the biosorbent, ranging from 0.43 to 89.66 mg/g, in the 12 trials run in duplicate. Regression analysis was performed on the results and the first-order polynomial equation was derived by representing the sorption of RO 13 dye as a function of the independent variables:
\( R^{2} = 99.94\% , R^{2} \left( {\text{pred}} \right) \, = 99.83\% , R^{2 } {\text{adj}} = \, 99.91\% \).
Analysis of the regression coefficients and the t values of six factors (Table 3) showed that the dye concentration (X 2) and contact time (X 6) had positive effect on biosorption, whereas pH (X 1), biosorbent dosage (X 3), temperature (X 4) and speed of agitation (X 5) had a negative effect on biosorption. The corresponding probability values (P values) indicate the significance of each of the coefficients, which in turn govern the patterns of interactions between the variables. The smaller the value of P, the more significant is the corresponding coefficient. Table 3 shows that all the parameters were significant (P < 0.001) in increasing the sorption by the biosorbent. The model was significant (P < 0.01) and R 2 = 0.9994 indicated that 99.94 % of the total variability in the response could be explained using this model.
The main effect plots of all the variables taken into consideration in this work were used to plot data means and also to compare magnitudes of marginal means as shown in Fig. 1.
A main effect occurs when the mean response changes across the levels of a factor. The points in the plot are the means of the response variables at various levels of each factor, with a reference line (center line) drawn across the plot representing the grand mean of the response data. A horizontal line (parallel to the x-axis) represents no main effect, while a non-horizontal line indicates a main effect. The steeper the slope of the line, the greater is the magnitude of the main effect. From Fig. 1, it is clear that though all the variables played an important role in the adsorption of RO 13 dye, pH, biosorbent dosage, dye concentration and speed of agitation seemed to be the dominant factors and were considered in the experiments of PSA and CCD. The grand mean of the response fell at SUC of around 35 mg/g.
The maximum removal of RO 13 dye was obtained at low pH (54.74 mg/g), while at higher pH the grand mean of SUC dropped down to 17.98 mg/g. Solution pH determines the surface charge of the adsorbent and the degree of ionization and speciation of the biosorbent, which affect the adsorption of dyes. Upon dissolution, ionic dyes release colored dye ions into the solution [25]. As the pH of the solution decreases, more protons are available to protonate the amino groups of chitosan molecules on the fungal cell wall to form positively charged –NH3 + groups. This increases electrostatic attraction between the anionic group (–SO3−) of the dye and the protonated amino group (–NH3 +) of chitosan, causing an increase in dye adsorption [9]. This chemical affinity between the positive charge on the biomass and negative charges in the structures of anionic dye RO 13 weakens the resistance of the boundary layer surrounding the adsorbent leading to more sorption under acidic conditions [25]. The deprotonation of surface groups in high pH range results in the electrostatic repulsion between the anionic dye and negatively charged sites, contributing to the decreased uptake of RO 13 dye in alkaline condition [26]. Furthermore, lower adsorption of the dye in alkaline medium is also due the competition from excess OH− ions with the anionic dye molecule for the adsorption sites [27].
It was seen that more RO 13 dye was adsorbed at higher dye concentration showing a positive effect on the system. The mean increased from 22.10 to 48.62 mg/g with an increase in dye concentration. Increased biosorption at higher concentration of RO 13 dye can be attributed to a combination of factors, i.e., increase in the driving force of the concentration gradient of the dye resulting in higher probability of collision between dye ions and the active sites on the biomass, and availability of functional groups on the surface of the biomass surrounded by a greater number of RO 13 ions leading to increased ability to bind dye ions [28–30]. Figure 1 also shows the adsorption of RO 13 as a function of biosorbent dosage. It was seen that increasing the biosorbent dosage in the system decreased the biosorption capacity, i.e., it had a negative effect on the system. The mean decreased from 47.37 to 23.35 mg/g when the biosorbent dosage was increased to +1 level. The increase in the biosorption at lower biosorbent dosage can be attributed to increased biosorbent surface area and the availability of more adsorption sites. Various reasons have been suggested to explain the reduced uptake capacity at higher biosorbent dosages such as competition of the ions for limited available sites, overlapping or aggregation of adsorption sites resulting in a decrease in the total adsorbent surface area, interference between binding sites and reduced mixing at higher biomass densities, and an increase in diffusion path length [31–35].
The uptake of RO13 dye appeared to be temperature dependent, showing maximum removal at lower temperature. This can be attributed to the decrease in the surface activity suggesting that adsorption between RO 13 dye on the dead biomass of R. arrhizus was an exothermic process [36]. A decrease in the biosorption of RO 13 dye at higher temperature can be related to the increasing Brownian movement of molecules in solution leading to the breaking of existing intermolecular hydrogen bonding between RO 13 dye and the biosorbent [37].
The speed of agitation showed a negative effect on the biosorption of RO 13 dye, decreasing the SUC from 43.09 mg/g at lower speed of agitation to 27.62 mg/g at higher speed of agitation. A decrease in the sorption capacity at higher speed of agitation may be attributed to improper contact between the dye ions and the binding sites on the biomass, as the suspension is no longer homogenous due to vortex formation, which makes the adsorption of dye ions difficult [38, 39]. The effect of contact time on the removal of RO 13 dye indicated that a longer contact time favored the reaction toward the equilibrium between the dye and the adsorbent. This may be due to the availability of the uncovered surface area on the biosorbent and the progressive occupation of active binding sites over a longer period of time.
The highest response of SUC of 89.66 mg/g after running Plackett–Burman experiments was obtained under the following conditions: pH 2.0; dye concentration 100 mg/L; biosorbent dosage 0.8 g/L; temperature 35 °C; speed of agitation 80 rpm; and contact time 120 min.
Locating the region of optimum response by the PSA
In the current investigation, PSA was employed to move from the current operating conditions to the optimum region in the most efficient way by using the minimum number of experiments. PSA was based on the zero level of the PBD and moved along the direction in which the dye concentration increased and pH, biosorbent dosage and speed of agitation decreased. The less significant factors, viz. temperature, was used in all trials at its −1 level (35 °C) for its negative contribution, while contact time was kept at its +1 level (120 min) for its positive contribution. The experimental design and results are shown in Table 4.
The highest response was found to be SUC of 124.02 mg/g with pH 2.0, dye concentration 103 mg/L, biosorbent dosage 0.8 g/L and speed of agitation 85 rpm. This point was concluded to be near the optimal point and was chosen for optimization by RSM using CCD.
Optimization of significant variables using CCD
CCD was employed at the specified combinations of four independent significant factors (pH, dye concentration, biosorbent dosage and speed of agitation) at five levels (−α, −1, 0, +1, +α) to study the interactions between them and to determine their optimum levels (Table 5). The levels of temperature and contact time were kept similar to the trial runs in PSA.
The design matrix of tested variables in 30 experimental runs along with the experimental results and the results of theoretically predicted responses (using the model equation) are shown in Table 6.
The SUC of the biomass increased to 133.63 mg/g after running the response surface design using the following conditions: pH 2.0, dye concentration 114 mg/L, biosorbent dosage 0.8 g/L and speed of agitation 85 rpm. Multiple regression analysis was used to analyze the data to obtain an empirical model for the best response and thus a second-order polynomial equation (Eq. 5) was derived as follows:
The mathematical expression of the relationships between the independent variables and dependent response is given in terms of uncoded factors. Apart from the linear effect of the parameter for the dye removal, the RSM also gives an insight into the quadratic and interaction effect of the parameters. These analyses are done by means of Fisher’s F test and Student’s t test. Student’s t test is used to determine the significance of the regression coefficients of the parameters. In general, the larger the magnitude of t and smaller the value of P, the more significant is the corresponding coefficient term [25]. The regression coefficient and the F and P values for all the linear, quadratic, and interaction effects of the parameters are given in Table 7.
From very small P values, it was observed that the coefficients for the linear, quadratic and interaction effects of the factors were highly significant except the quadratic effect for dye concentration (P = 0.550), interaction effects for pH and dye concentration (P = 0.314), and pH and biosorbent dosage (P = 0.514). These measures indicated that the accuracy and general ability of the polynomial model were good and that analysis of the response trends using the model was reasonable. The statistical significance of the ratio of the mean square variation due to the regression and mean square residual error was also tested using analysis of variance (ANOVA) as shown in Table 8.
ANOVA is a statistical technique that subdivides the total variation in a set of data into component parts associated with specific sources of variation for the purpose of testing hypothesis on the parameters of the model. The ANOVA of the quadratic regression model demonstrated that the model was highly significant, as was evident from the low P value of the Fisher’s F test (F model, 32.90) [(P model > F) = 0.000]. The coefficient of variation is the error expressed as a percentage of the mean. For the proposed model to be a good predictor of the experimental results, the F model value should be greater than the calculated value of the F distribution for a certain number of degrees of freedom in the model at the level of significance α. The lower calculated F 14,14 value (8.99) than the tabulated F value even at the 0.0001 confidence level showed a statistically insignificant lack of fit. The model was found to be adequate for prediction within the range of variables employed. The coefficient of determination R 2 = 0.9770 implied a good agreement between the experimental and predicted values of biosorption, which can be attributed to the given independent variables. R 2 also indicated that only 2.30 % of the total variations could not be explained by the model. The pred-R 2 of 0.94 was in reasonable agreement with adj-R 2 of 0.93, indicating that the model was significant. The adjusted coefficient of determination represents the proportion of the variation in the response explained by the regression model. It is thus envisaged that Eq. (5) can capture 93 % of the variation in the measured values of SUC as function of the four independent conditions within the ranges considered in the present study. The coefficient of variation is the error expressed as a percentage of the mean. It indicates the degree of precision with which the treatments are compared. The value of CV (2.46 %) demonstrated that the performed experiments were reliable.“Adeq Precision” measures the signal to noise ratio. A ratio greater than 4 is desirable. A ratio of 17.602 obtained in this study indicated an adequate signal. Therefore, this model can be used to navigate the design space. The ANOVA thus indicated that the second-order polynomial model for Eq. 4 was highly significant and adequate to represent the actual relationship between the response (SUC mg/g) and variables, with P < 0.000 and a high value of the coefficient of determination (97.7 %).
The graphical representations of the regression model, called the 3D surface plots, were obtained to explain the interaction of variables and to determine the optimum level of each variable (pH, dye concentration, biosorbent dosage and speed of agitation) for maximum response (SUC) (Fig. 2a–d).
A surface plot can be used to explore the relationship between three variables. Here, each response surface plot represented the effect of two independent variables, holding the other variables at zero level. These surface plots provide a method to predict the biosorption efficiency for different values of the tested variables. As seen in Fig. 1a–d, each response surface yield showed a clear peak suggesting that the optimum point was inside the design boundary. The surface plot was based on independent variables, i.e., dye concentration (X 2) and biosorbent dosage (X 3), while the other independent variables pH (X 1) and speed of agitation (X 5) were kept at zero level as shown in Fig. 2a. An increase in SUC could be achieved when the dye concentration increased from 97.5 to 108.5 mg/L, and beyond this SUC decreased sharply even for a tiny increase in dye concentration. The SUC increased when the biosorbent dosage decreased from 0.8 to 0.73 g/L. The profile observed in Fig. 2b with speed of agitation (X 5) and biosorbent dosage (X 3) suggests that the SUC of the system increased with increasing biosorbent dosage, while change in speed of agitation showed very little effect on SUC. The two-dimensional contour plot showed a clearly elongated line running diagonally on the plot, suggesting that the interaction between speed of agitation (X 5) and biosorbent dosage (X 3) was significant on SUC. The response surface plot shown in Fig. 2c suggested an increase in SUC with an increase in speed of agitation from 85 to 87.3 rpm, and dye concentration from 103 to 108.5 mg/L. The effect of pH (X 1) and agitation speed (X 5) on biosorption while keeping dye concentration (X 2) and biosorbent dosage (X 3) constant is depicted in Fig. 2d. A slight increase in SUC could be achieved with increase in speed of agitation from 85 to 87.3 rpm and decrease in pH from 2.0 to 1.78.
Validation of the experimental model
The special feature of the RSM tool, ‘point prediction’, was employed to find the optimum value of the combination of the four factors for maximum biosorption. The predicted optimal conditions were as follows: pH 1.78, dye concentration 108.5 mg/L, biosorbent dosage 0.73 g/L and speed of agitation 87.3 rpm. Validation experiments were performed to verify the accuracy of the model. The results showed that the predicted values were in accordance with the experimental results. The biosorption was 133.0 mg/g, which was in excellent agreement with the predicted value (133.97 mg/g). The result showed good correlation between statistically predicted and experimental values, which confirmed the model’s authenticity and the existence of an optimum point. Therefore, response surface optimization could be successfully used to optimize the process parameters and evaluate the performance of R. arrhizus as a biosorbent and to achieve an overall 49.04 % increase in biosorption.
Artificial neural network (ANN)
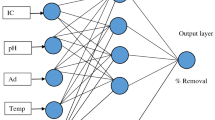
JMP (version 10) computing environment was chosen to generate a neural network to test and validate the model. ANNs are inspired by biological neural systems. In this approach, weighted sum of inputs arriving at each neuron is passed through an activation function (generally nonlinear) to generate an output signal. Each input node represents an independent variable, while the output nodes give the dependent variables [32]. A fully connected holdback network with the input, hidden and output layers is shown in the Fig. 3.
Input layer and hidden layer have nonlinear activation neurons, and the output layer has linear neurons in network topology [22]. Each node in the input layer represents the value of one independent variable, while the output nodes indicate the dependent variables [38]. The model consisted of three layers: an input layer with four neurons (initial pH, dye concentration, biosorbent dosage and speed of agitation), a hidden layer with three neurons and an output with one neuron (4-3-1). Figure 4 shows a comparison between experimental and predicted values of SUC using the neural network model. From Fig. 4, it was observed that the simulated result using ANN showed a satisfactory correlation with the experimental result. The trained network gave a mean square error (MSE) of 0.02 with correlation coefficient (R 2) of 0.83, indicating that the ANN model reproduced the adsorption well in this system.
Conclusion
A low-cost biosorbent prepared from dead fungal biomass of R. arrhizus NCIM 997 was successfully applied for the removal of RO 13 dye, a reactive mono-azo dye from aqueous solution using sequential optimization strategy. The statistical approach consisting of Plackett–Burman design, path of steepest ascent and central composite design helped in rapidly identifying important factors, studying the interactions between them and determining their optimum levels. Development of a polynomial model showed significant results for optimizing biosorption conditions for the RO 13 dye. This study clearly showed that the RSM is an effective tool for optimization of process parameters to maximize the dye removal. The optimum values of pH, dye concentration, biosorbent dosage and speed of agitation were found to be 2.0, 0.8 g/L, 114.0 mg/L and 85 rpm for the removal of RO 13 dye, respectively. The experimental values were in good agreement with the model-predicted values. ANN results showed that neural network modeling could effectively simulate and predict the behavior of the process. Optimal conditions obtained in this experiment have laid a solid foundation for further use of this microorganism in the treatment of reactive azo dyes containing effluents.
References
Shin M, Nguyen T, Ramsay J (2002) Evaluation of support materials for the surface immobilization and decoloration of amaranth by Trametes versicolor. App J Microbiol Biotechnol 60:218–223
Asamudo NU, Daba AS, Ezeronye OU (2005) Bioremediation of textile effluent using Phanerochaete chrysosporium. Afr J Biotechnol 4(13):1548–1553
Zille A (2005) Laccase reactions for textile applications. Dissertation, Universidade do Minho, Escola de Engenharia
Sadaf S, Bhatti HN (2014) Evaluation of peanut husk as a novel, low cost biosorbent for the removal of Indosol Orange RSN dye from aqueous solutions: batch and fixed bed studies. Clean Techn Environ Policy 16(3):527–544
Sadaf S, Bhatti HN, Nausheen S, Noreen S (2014) Potential use of low-cost lignocellulosic waste for the removal of Direct Violet 51 from aqueous solution: equilibrium and breakthrough studies. Arch Environ Contam Toxicol 66(4):557–571
Nawaza S, Bhatti HN, Bokharia TH, Sadaf S (2014) Removal of Novacron Golden Yellow dye from aqueous solutions by low-cost agricultural waste: Batch and fixed bed study. Chem Ecol 30(1):52–65
Noreen S, Bhatti HN, Nausheen S, Sadaf S, Ashfaq M (2013) Batch and fixed bed adsorption study for the removal of Drimarine Black CL-B dye from aqueous solution using a lignocellulosic waste: a cost affective adsorbent. Ind Crop Prod 50:568–579
Das N, Vimala R, Karthika P (2008) Biosorption of heavy metals—an overview. Ind J Biotechnol 7:159–169
Chen AH, Chen SM (2009) Biosorption of azo dyes from aqueous solution by glutaraldehyde-crosslinked chitosans. J Hazard Mater 172:1111–1121
Vijayaraghavan K, Yun Y (2008) Polysulfone-immobilized Corynebacterium glutamicum: a biosorbent for Reactive black 5 from aqueous solution in an up-flow packed column. Chem Eng J145:44–49
Ambrósio ST, José C, Vilar J, Carlos A, Alves S, Kaoru O, Nascimento AE, Longo RL, Takaki GMC (2012) A Biosorption isotherm model for the removal of reactive azo dyes by inactivated mycelia of Cunninghamella elegans UCP542. Molecules 17:452–462
de Sales PF, Magriotis ZM, Rossi M, Resende RF, Nunes CA (2013) Optimization by response surface methodology of the adsorption of Coomassie Blue dye on natural and acid-treated clays. J Environ Manag 130:417–428
Mourabet M, ElRhilassi A, Bennani-Ziatni M, Taitai A (2014) Comparative study of Artificial Neural Network and Response Surface Methodology for modelling and optimization the adsorption capacity of fluoride onto apatitictricalcium phosphate. Univ J Appl Math 2(2):84–91
Aber S, Amani-GhadimA R, Mirzajani V (2009) Removal of Cr(VI) from polluted solutions by electrocoagulation: modeling of experimental results using artificial neural network. J Hazard Mater 171:484–490
Rosa JM, Prado KRM, Alves WAL, Pereira FH, Santana JCC, Tambourgi EB (2013) Applying of a Neural Network in effluent treatment simulation as an environmental solution for textile industry. Chem Eng Trans 32:73–78
Aleboyeh A, Daneshvar N, Kasiri MB (2007) Optimization of C.I. Acid Red 14 azo dye removal by electrocoagulation batch process with Response Surface Methodology. Chem Eng Proc 47:827–832. doi:10.1016/j.cep.2007.01.033
Prakash N, Manikandan SA, Govindarajan L, Vijayagopal V (2008) Prediction of biosorption efficiency for the removal of copper(II) using artificial neural networks. J Hazard Mater 152:1268–1275
Pareek VK, Brungs MP, Adesina AA, Sharma R (2002) Artificial neural network modeling of a multiphase photodegradation system. J Photochem Photobiol A Chem 149:139–146
O’Mahony T, Guibal E, Tobin JM (2002) Reactive dye biosorption by Rhizopus arrhizus biomass. Enz Microb Technol 31:456–463
Gül ÜD, Silah H (2014) Comparison of color removal from reactive dye contaminated water by systems containing fungal biosorbent, active carbon and their mixture. Water Sci Technol. doi:10.2166/wst.2014.339.Science
Machado KMG, Compart LCA, Morais RRO, Santos MH (2006) Biodegradation of reactive textile dyes by basidiomycetous fungi from Brazilian ecosystems. Braz J Microbiol 37:481–487
Tunali S, Kiran I, Akar T (2005) Chromium (VI) biosorption characteristics of Neurospora crassa fungal biomass. Miner Eng 18:681–689
Plackett RL, Burman JP (1946) The design of optimum multifactorial experiments. Biometrika 33:305–325
Zhao D-H, Zhang Y-L, Wei Y-P, Gao H-W (2009) Facile eco-friendly treatment of a dye wastewater mixture by in situ hybridization with growing calcium carbonate. J Mater Chem 19:7239–7244
Montgomery DC (1991) Design of Experiments. In: Response surface methodology, 3rd edn. Wiley, New York
Ramakrishna KR, Viraraghavan T (1997) Use of slag for dye removal. Waste Manag 17:483–488
Wong SY, Tan YP, Abdullah AH, Ong ST (2009) Removal of Basic Blue 3 and Reactive Orange 16 by adsorption onto quartenized sugar cane bagasse. Malaysian J Anal Sci 13(2):185–193
Uzun I (2006) Kinetics of the adsorption of reactive dyes by chitosan. Dyes Pigments 70:76–83. doi:10.1016/j.dyepig.2005.04.016
Aksakal O, Ucun H (2010) Equilibrium, kinetic and thermodynamic studies of the biosorption of textile dye (Reactive Red 195) onto Pinus sylvestris L. J Hazard Mater 181(1–3):666–672
Ho YS, McKay G (2000) Correlative biosorption equilibria model for a binary batch system. Chem Eng Sci 55:817–825
Baral A, Engel ken RD (2002) Chromium-based regulations and greening in metal finishing industries in the USA. Environ Sci Policy 5:121–133
Gong R, Sun Y, Chen J, Liu H, Yang C (2005) Effect of chemical modification on dye adsorption capacity of peanut hull. Dyes Pigments 67:175–181. doi:10.1016/j.dyepig.2004.12.003
Jaikumar V (2009) Effect of Biosorption parameters kinetics isotherm and thermodynamics for acid green dye biosorption from aqueous solution by brewery waste. Int J Chem 1(1):21–29
Kahraman S, Asma D, Erdemoglu S, Yesilada O (2005) Biosorption of copper by live and dried biomass of Phanerocheate crysoporium and Funalia trogii. Eng Life Sci5 (1): 72–77
Vijayaraghavan K, Yun Y (2008) Polysulfone-immobilized Corynebacterium glutamicum: a biosorbent for Reactive black 5 from aqueous solution in an up-flow packed column. Chem Eng J 145:44–49
Ponnusamy SK, Subramaniam R (2013) Process optimization studies of Congo red dye adsorption onto cashew nut shell using Response Surface Methodology. Int J Ind Chem 4:17. http://www.industchem.com/content/4/1/17
Li H, Huang G, An C, Hu J, Yang S (2013) Removal of tannin from aqueous solution by adsorption onto treated coal fly ash: kinetic, equilibrium, and thermodynamic studies. Ind Eng Chem Res 52:15923–15931
Ajay Kumar AV, Darwish NA, Hilal N (2009) Study of various parameters in the biosorption of heavy metals on activated sludge. World Appl Sci 5:32–40
Bhatti MS, Kapoor D, Kalia RK, Reddy AS, Thukral AK (2011) RSM and ANN modeling for electro-coagulation of copper from simulated wastewater: multi objective optimization using genetic algorithm approach. Desalin Technol 274:74–80
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Saraf, S., Vaidya, V.K. Statistical optimization of biosorption of Reactive Orange 13 by dead biomass of Rhizopus arrhizus NCIM 997 using response surface methodology. Int J Ind Chem 6, 93–104 (2015). https://doi.org/10.1007/s40090-015-0036-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40090-015-0036-x