Abstract
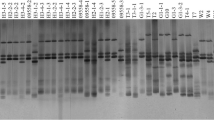
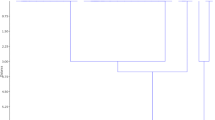
Bacterial blight (BB), caused by Xanthomonas oryzae pv. oryzae (Xoo), is a serious constraint to basmati rice production. The increasingly evident outbreaks of BB in recent years necessitate an immediate search for ecology-conscious, cost-effective and durable management strategy. Given the context, this investigation aimed to generate knowledge about the haplotypic and pathogenic variability of Xoo population severely affecting Pusa Rice Hybrid 10 (PRH-10), world’s first superfine aromatic basmati hybrid. Seven haplotypes were detected from the experimental field applying two PCR-based assays, i.e. repetitive sequence-based polymerase chain reaction and insertion sequence based PCR. The experimental results revealed greater genetic heterogeneity in the Xoo population with high value of total haplotypic diversity (H T = 0.79). In addition, based on the virulence assay, the genetic heterogeneity corresponded to the presence of four pathotypes. Importantly, the substantial line × strain interactions was significant (p ≤ 0.01), which confirmed the host specificity in the system. In parallel, it was noted that most of the key resistance genes except Xa21, deployed in the rice breeding program at International Rice Research Institute were overcome by the Xoo strains, thus underscoring the relevance of this gene in resistance breeding. Therefore, incorporating Xa21 alone, or in combination with xa13 or xa5 into the parental lines, using various modern genomics-assisted approaches might be the potential fast-track strategy to breed PRH-10 with remarkably enhanced BB resistance. The present investigation provides a molecular framework for future epidemiological studies and is likely to assist rice breeders to expedite basmati rice improvement.




Similar content being viewed by others
References
Mew TW (1987) Current status and future prospects of research on bacterial blight of rice. Annu Rev Phytopathol 25:359–382
Pandey MK, Rani NS, Sundaram RM, Laha GS, Madhav MS, Rao KS, Sudharshan I, Hari Y, Varaprasad GS, Rao LVS, Suneetha K, Sivaranjani AKP, Viraktamath BC (2013) Improvement of two traditional Basmati rice varieties for bacterial blight resistance and plant stature through morphological and marker-assisted selection. Mol Breeding 31:239–246
Pandey S, Singh B, Kumar J (2014) DNA typing and virulence determination of Xanthomonas oryzae pv. oryzae population for the management of bacterial leaf blight of rice in Udham Singh Nagar, India. Eur J Plant Pathol 138:847–862
Verdier V, Vera Cruz C, Leach JE (2011) Controlling rice bacterial blight in Africa: needs and prospects. J Biotechnol. doi:10.1016/j.jbiotec.2011.09.020
Collard BCY, Mackill DJ (2008) Marker assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363:557–572
Sundaram RM, Vishnupriya MR, Biradar SK, Laha GS, Shobharani N, Sharma MP, Sonti RV (2008) Marker assisted introgression of bacterial blight resistance in Sambha Mahsuri, an elite indica rice variety. Euphytica 160:411–422
Sundaram RM, Vishnupriya MR, Laha GS, Rani NS, Rao PS, Balachandran SM, Reddy GA, Sarma NP, Sonti RV (2009) Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety. Biotechnol J 4:400–407
Tan GX, Ren X, Weng QM, Shi ZY, Zhu LL, He GC (2004) Mapping of a new resistance gene to bacterial blight in rice line introgressed from Oryza officinalis. Yi Chuan Xue Bao 31:724–729 (in Chinese)
Luo Y, Zakaria S, Basyah B, Ma T, Li Z, Yang J Yin Z (2014) Marker-assisted breeding of Indonesia local rice variety Siputeh for semi-dwarf phenotype, good grain quality and disease resistance to bacterial blight. Rice 7–33
Vera Cruz CM, Bai JF, Ona I, Leung H, Nelson RJ, Mew TW, Leach JE (2000) Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc Natl Acad Sci USA 97:13500–13505
Leung H, Nelson RJ, Leach JE (1993) Population structure of plant pathogenic fungi and bacteria. Adv Plant Pathol 10:157–250
Adhikari TB, Basnayat RC, Mew TW (1999) Virulence of Xanthomonas oryzae pv. oryzae on rice lines containing single resistance genes and gene combinations. Plant Dis 83:46–50
Adhikari TB, Mew TW, Teng PS (1994) Phenotypic diversity of Xanthomonas oryzae pv. oryzae in Nepal. Plant Dis 78:68–72
Adhikari TB, VeraCruz CM, Zhang Q, Nelson RJ, Skinner DZ, Mew TW, Leach JE (1995) Genetic diversity of Xanthomonas oryzae pv. oryzae in Asia. Appl Environ Microbiol 61:966–971
Ardales EY, Leung H, Vera Cruz CM, Mew TW, Leach JE, Nelson RJ (1996) Hierarchical analysis of spatial variation of the rice bacterial blight pathogen across diverse agroecosystems in the Philippines. Phytopathology 86:241–252
Finckh MR, Nelson RJ (1999) Phylogenetic and pathotypic analysis of rice babacterial blight Race 3. Eur J Plant Pathol 105:743–751
Leach JE, Rhoads ML, Vera Cruz CM, White FF, Mew TW, Leung H (1992) Assessment of genetic diversity and population structure of Xanthomonas oryzae pv. oryzae within a repetitive DNA element. Appl Environ Microbiol 58:2188–2195
Lore JS, Vikal Y, Hunjan MS, Goel RK, Bharaj TS, Raina TL (2010) Genotypic and pathotypic diversity of Xanthomonas oryzae pv. oryzae, the cause of bacterial blight of rice in Punjab state of India. J Phytopathol 1–9
Noda T, Li C, Li J, Ochiai H, Ise K, Kaku H (2001) Pathogenic diversity of Xanthomonas oryzae pv. oryzae strains from Yunnan province, China. JARQ 35:97–103
Shanty ML, George MLC, Vera Cruz CM, Bernardo MA, Nelson RJ, Leung H, Reddy JN, Sridhar R (2001) Identification of resistance genes effective against rice bacterial blight pathogen in Eastern India. Plant Dis 85:506–512
Vera Cruz CM, Ardales EY, Skinner DZ, Talag J, Nelson RJ, Louws FJ, Leung H, Mew TW, Leach JE (1996) Measurement of haplotypic variation in Xanthomonas oryzae pv. oryzae within a single field by rep-PCR and RFLP analysis. Phytopathology 86:1352–1359
Adhikari TB, Mew TW, Leach JE (1999) Genotypic and pathotypic diversity in Xanthomonas oryzae pv.oryzae in Nepal. Phytopathology 89:687–694
De Bruijn FL (1992) Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti strains and other soil bacteria. Appl Environ Microbiol 58:2180–2187
Dudd AK, Sadowsky MJ, De Bruijn FJ (1993) Use of repetitive sequences and the polymerase chain reaction to classify genetically related Bradyrhizibium japonicum serocluster 123 strains. Appl Environ Microbiol 59:1702–1708
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in Eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:406–409
Gupta VS, Rajebhosale MD, Sodhi M, Singh S, Gnanamanickam SS, Dhaliwal HS (2001) Assessment of genetic variability and strain identification of Xanthomonas oryzae pv. oryzae using RAPD-PCR and IS1112-based PCR. Curr Sci 80(8):1043–1048
Nino-Liu DO, Ronald PC, Bogdanove AJ (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol 7(5):303–324
Mew TW, Vera Cruz CM, Medalla ES (1992) Changes in race frequency of Xanthomonas oryzae pv. oryzae in response to rice cultivars planted in the Philippines. Plant Dis 76:1029–1032
Basavaraj SH, Singh VK, Singh A, Singh A, Singh A, Singh A, Anand D, Yadav S, Ellur RK, Singh D, Krishnan SJ, Nagarajan M, Mohapatra T, Prabhu KV, Singh AK (2010) Marker assisted improvement of bacterial blight resistance in parental lines of Pusa RH 10, a superfine grain aromatic rice hybrid. Mol Breeding 26:293–305
Ntsys PC version 2.11 s; Rohlf FJ (2000) NTSYS-pc numerical taxonomy and multivariate analysis system version 2.1. Exeter Software, Setauket, New York, USA
Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification. Freeman, San Francisco
Yap IV, Nelson RJ (1996) WinBoot: a program for performing bootstrap analysis of binary data to determine the confidence limits of UPGMA-based dendrograms. IRRI Disc Series No. 14. International Rice Research Institute, Manila, Philippines
Nei M (1973) Analysis of gene diversity in subdivided population. Proc Natl Acad Sci USA 70(12):3321–3323
Kauffman HE, Reddy APK, Hsieh SPY, Merca SD (1973) An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep 57:537–541
Bolger ME, Weisshaar B, Scholz U, Stein N, Usadel B, Mayer KF (2014) Plant genome sequencing-applications for crop improvement. Curr Opin Biotechnol 26:31–37
Mishra D, Vishnupriya RM, Anil MG, Kondal K, Raj Y, Sonti RV (2013) Pathotype and genetic diversity amongst Indian isolates of Xanthomonas oryzae pv. oryzae. PLoS ONE 8(11):1–11
Wang GL, Song WY, Ruan DL, Sideris S, Ronald PC (1996) The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae strains in transgenic plants. Mol Plant Microbe Interact 9:850–855
Devadath S (1983) A strain of Oryzae barthii an African wild rice immune to bacterial blight of rice. Curr Sci 52:27–28
Zhai W, Wang W, Zhou Y, Li X, Zhang Q, Wang G (2002) Breeding bacterial blight-resistant hybrid rice with the cloned bacterial blight resistance gene Xa21. Mol Breeding 8:285–293
Acknowledgments
The authors are grateful to International Rice Research Institute, Manila, Philippines for providing the seeds of near-isogenic lines. They also thank the reviewers for critical reading and valuable suggestions regarding the manuscript. The authors declare that there are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pandey, S., Bohra, A., Singh, B. et al. Haplotypic Diversity and Virulence of Xanthomonas oryzae pv. oryzae Population Infecting the First Superfine Aromatic Basmati Hybrid. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 87, 1005–1014 (2017). https://doi.org/10.1007/s40011-015-0675-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-015-0675-x