Abstract
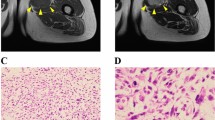
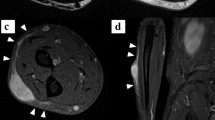
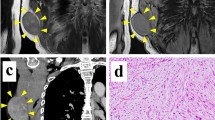
Myxofibrosarcoma (MFS) is among the most aggressive and complex sarcoma types that require novel therapeutic approaches for improved clinical outcomes. MFS displays highly complex karyotypes, and frequent alterations in p53 signaling and cell cycle checkpoint genes as well as loss-of-function mutations in NF1 and PTEN have been reported. The effects of radiotherapy and chemotherapy on MFS are limited, and complete surgical resection is the only curative treatment. Thus, the development of novel therapeutic strategies for MFS has long been long desired for MFS. Patient-derived cell lines are an essential tool for basic and translational research in oncology. However, public cell banks provide only a limited number of MFS cell lines. In this study, we aimed to develop a novel patient-derived MFS cell line, which was established from the primary tumor tissue of a 71-year-old male patient with MFS and was named NCC-MFS2-C1. A single-nucleotide polymorphism assay revealed that NCC-MFS2-C1 cells exhibited gain and loss of genetic loci. NCC-MFS2-C1 cells were maintained as a monolayer culture for over 24 passages for 10 months. The cells exhibited spindle-like morphology, continuous growth, and capacity for spheroid formation and invasion. Screening of 213 anticancer agents revealed that bortezomib, gemcitabine, romidepsin, and topotecan at low concentrations inhibited the proliferation of NCC-MFS2-C1 cells. In conclusion, we established a novel MFS cell line, NCC-MFS2-C1, which can be used for studying the molecular mechanisms underlying tumor development and for the in vitro screening of anti-cancer drugs.




Similar content being viewed by others
References
Fletcher CDM, Bridge JA, Hogendoorn P, Mertens F. Soft tissue and bone tumours. 4th ed. Geneva: WHO Press; 2020.
Ogura K, Hosoda F, Arai Y, et al. Integrated genetic and epigenetic analysis of myxofibrosarcoma. Nat Commun. 2018;9:2765.
Barretina J, Taylor BS, Banerji S, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–21.
Haglund KE, Raut CP, Nascimento AF, Wang Q, George S, Baldini EH. Recurrence patterns and survival for patients with intermediate- and high-grade myxofibrosarcoma. Int J Radiat Oncol Biol Phys. 2012;82:361–7.
Sanfilippo R, Miceli R, Grosso F, et al. Myxofibrosarcoma: prognostic factors and survival in a series of patients treated at a single institution. Ann Surg Oncol. 2011;18:720–5.
Mentzel T, Calonje E, Wadden C, et al. Myxofibrosarcoma. Clinicopathologic analysis of 75 cases with emphasis on the low-grade variant. Am J Surg Pathol. 1996;20:391–405.
Merck C, Angervall L, Kindblom LG, Oden A. Myxofibrosarcoma. A malignant soft tissue tumor of fibroblastic-histiocytic origin. A clinicopathologic and prognostic study of 110 cases using multivariate analysis. Acta pathologica, microbiologica, et immunologica Scandinavica Supplement. 1983;282:1–40.
Weiss SW, Enzinger FM. Myxoid variant of malignant fibrous histiocytoma. Cancer. 1977;39:1672–85.
Willems SM, Debiec-Rychter M, Szuhai K, Hogendoorn PC, Sciot R. Local recurrence of myxofibrosarcoma is associated with increase in tumour grade and cytogenetic aberrations, suggesting a multistep tumour progression model. Mod Pathol. 2006;19:407–16.
Lin CN, Chou SC, Li CF, et al. Prognostic factors of myxofibrosarcomas: implications of margin status, tumor necrosis, and mitotic rate on survival. J Surg Oncol. 2006;93:294–303.
Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5.
Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–61.
Seashore-Ludlow B, Rees MG, Cheah JH, et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5:1210–23.
Rees MG, Seashore-Ludlow B, Cheah JH, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12:109–16.
Haverty PM, Lin E, Tan J, et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature. 2016;533:333–7.
Iorio F, Knijnenburg TA, Vis DJ, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–54.
Behan FM, Iorio F, Picco G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–6.
Townsend EC, Murakami MA, Christodoulou A, et al. The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell. 2016;29:574–86.
Kawashima H, Ogose A, Gu W, et al. Establishment and characterization of a novel myxofibrosarcoma cell line. Cancer Genet Cytogenet. 2005;161:28–35.
Huang HY, Wu WR, Wang YH, et al. ASS1 as a novel tumor suppressor gene in myxofibrosarcomas: aberrant loss via epigenetic DNA methylation confers aggressive phenotypes, negative prognostic impact, and therapeutic relevance. Clin Cancer Res. 2013;19:2861–72.
Lohberger B, Stuendl N, Wolf E, Liegl-Atzwanger B, Leithner A, Rinner B. The novel myxofibrosarcoma cell line MUG-Myx1 expresses a tumourigenic stem-like cell population with high aldehyde dehydrogenase 1 activity. BMC Cancer. 2013;13:563.
Salawu A, Fernando M, Hughes D, et al. Establishment and molecular characterisation of seven novel soft-tissue sarcoma cell lines. Br J Cancer. 2016;115:1058–68.
Lohberger B, Stuendl N, Leithner A, et al. Establishment of a novel cellular model for myxofibrosarcoma heterogeneity. Sci Rep. 2017;7:44700.
Ariizumi T, Ogose A, Kawashima H, Hotta T, Umezu H, Endo N. Multinucleation followed by an acytokinetic cell division in myxofibrosarcoma with giant cell proliferation. J Exp Clin Cancer Res. 2009;28:44.
Kito F, Oyama R, Sakumoto M, et al. Establishment and characterization of a novel cell line, NCC-MFS1-C1, derived from a patient with myxofibrosarcoma. Hum Cell. 2019;32:214–22.
Yoshimatsu Y, Noguchi R, Tsuchiya R, et al. Establishment and characterization of NCC-CDS2-C1: a novel patient-derived cell line of CIC-DUX4 sarcoma. Hum Cell. 2020;33:427–36.
Workgroup ATCCSDO. Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10:441–8.
Capes-Davis A, Reid YA, Kline MC, et al. Match criteria for human cell line authentication: where do we draw the line? Int J Cancer. 2013;132:2510–9.
Capes-Davis A, Dirks W, MacLeod R, Uphoff C. Quality Matters: cell lines and their use in research. GIT Lab J Eur. 2014;17:12–3.
Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72.
Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63.
Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21:4084–91.
Forbes SA, Tang G, Bindal N, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657657.
Hattori E, Oyama R, Kondo T. Systematic review of the current status of human sarcoma cell lines. Cells. 2019;8:157.
Moneo V, Serelde BG, Fominaya J, et al. Extreme sensitivity to Yondelis (Trabectedin, ET-743) in low passaged sarcoma cell lines correlates with mutated p53. J Cell Biochem. 2007;100:339–48.
Miserocchi G, De Vita A, Mercatali L, et al. Characterization and drug sensitivity of a new high-grade myxofibrosarcoma cell line. Cells. 2018;7:186.
Maki RG, Kraft AS, Scheu K, et al. A multicenter Phase II study of bortezomib in recurrent or metastatic sarcomas. Cancer. 2005;103:1431–8.
Acknowledgements
We appreciate the technical support by Miss Yu Kuwata (Division of Rare Cancer Research, National Cancer Center). We would also like to thank the Tochigi Cancer Center Operating Room Nurse Team and the Secretary of the Medical Office for their assistance in processing and transporting the samples. The Fundamental Innovative Oncology Core at the National Cancer Center provided support for the SNParray experiment. We would like to thank Editage (www.editage.jp) for English editing and constructive comments on the manuscript.
Funding
This research was supported by the Japan Agency for Medical Research and Development grant 20ck0106537h0001, “Study to Overcome the Limits of Cancer Genome-based Medicine Using Patient-derived “Rare Cancer” Model”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the ethics committee of the Tochigi Cancer Center and the National Cancer Center, and written informed consent was obtained from the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13577_2020_420_MOESM3_ESM.tif
Supplementary Figure 3 Growth curves with agents that showed relatively higher inhibitory effects on NCC-MFS2-C1 cell proliferation (TIF 689 kb)
13577_2020_420_MOESM6_ESM.xlsx
Supplementary Table 3 IC50 values of four agents that showed high inhibitory effects on NCC-MFS2-C1 cell proliferation (XLSX 11 kb)
Rights and permissions
About this article
Cite this article
Noguchi, R., Yoshimatsu, Y., Ono, T. et al. Establishment and characterization of NCC-MFS2-C1: a novel patient-derived cancer cell line of myxofibrosarcoma. Human Cell 34, 246–253 (2021). https://doi.org/10.1007/s13577-020-00420-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-020-00420-z