Abstract
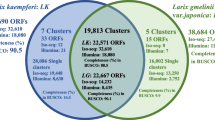
Larix kaempferi is widely cultivated because of its significant ecological and economic values. However, the molecular mechanism underlying the growth and development of Larix kaempferi remains largely unknown because of its long growth cycle and highly complex genetic background. In this study, global transcriptome sequencing of Larix kaempferi was conducted via Solexa sequencing. In total, 15.49 million clean reads were assembled into 20,395 unigenes and then annotated. The study found hundreds of highly expressed unigenes involved in growth and development (e.g. plant hormone regulation pathways or starch and sucrose metabolic pathways), of which 25 unigenes with full-length coding regions interested us the most. In addition, two gene families containing the zinc finger and nucleotide-binding site (NBS) domains were further explored. A total of 48 C3H-type zinc finger genes, most containing a conversed C-X7–10-C-X5-C-X3-H motif, were identified and divided into eight distinct subfamilies through phylogenetic analysis. Twenty-one genes containing the NBS domain represented four subfamilies. These findings provided new insights into the transcriptome characteristics of Larix kaempferi and abundant sequence information to elucidate the roles of the functional genes in Larix kaempferi.




Similar content being viewed by others
References
Arnaud D, Dejardin A, Leple JC, Lesage-Descauses MC, Pilate G (2007) Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res 14:103–116
Chai G, Hu R, Zhang D, Qi G, Zuo R, Cao Y, Chen P, Kong Y, Zhou G (2012) Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genom 13:253
Chen Y, Sun A, Wang M, Zhu Z, Ouwerkerk PB (2014) Functions of the CCCH type zinc finger protein OsGZF1 in regulation of the seed storage protein GluB-1 from rice. Plant Mol Biol 84:621–634
Chou ML, Shih MC, Chan MT, Liao SY, Hsu CT, Haung YT, Chen JJ, Liao DC, Wu FH, Lin CS (2013) Global transcriptome analysis and identification of a CONSTANS-like gene family in the orchid Erycina pusilla. Planta 237:1425–1441
Fernández M, Troncoso V, Valenzuela S (2015) Transcriptome profile in response to frost tolerance in Eucalyptus globules. Plant Mol Biol Rep 33:1472–1485
Gordo SM, Pinheiro DG, Moreira EC, Rodrigues SM, Poltronieri MC, de Lemos OF, da Silva IT, Ramos RT, Silva A, Schneider H, Silva WA Jr, Sampaio I, Darnet S (2012) High-throughput sequencing of black pepper root transcriptome. BMC Plant Biol 12:168
Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, Shinozaki K, Nakashima K, Yamaguchi-Shinozaki K (2013) OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol 161:1202–1216
Jin Z, Xu W, Liu A (2014) Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta 239:299–312
Kang YJ, Kim KH, Shim S, Yoon MY, Sun S, Kim MY, Van K, Lee SH (2012) Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol 12:139
Kramer S, Kimblin NC, Carrington M (2010) Genome-wide in silico screen for CCCH-type zinc finger proteins of Trypanosoma brucei, Trypanosoma cruzi and Leishmania major. BMC Genom 11:283
Krishna SS, Majumdar I, Grishin NV (2003) Structural classification of zinc fingers. Nucleic Acids Res 31:532–550
Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, Reidel EJ, Turgeon R, Liu P, Sun Q, Nelson T, Brutnell TP (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42:1060–1067
Li S, Zhao B, Yuan D, Duan M, Qian Q, Tang L, Wang B, Liu X, Zhang J, Wang J, Sun J, Liu Z, Feng YQ, Yuan L, Li C (2013a) Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc Natl Acad Sci USA 110:3167–3172
Li A, Zhou YN, Jin C, Song WQ, Chen CB, Wang CG (2013b) LaAP2L1, a heterosis- associated AP2/EREBP transcript factor of Larix, is sufficient to increase organ size and final biomass by effecting cell proliferation in Arabidopsis. Plant Cell Physiol 54:1822–1836
Li SG, Li WF, Han SY, Yang WH, Qi LW (2013c) Stage-specific regulation of four HD-ZIP III transcription factors during polar pattern formation in Larix leptolepis somatic embryos. Gene 522:177–183
Lin PC, Pomeranz MC, Jikumaru Y, Kang SG, Hah C, Fujioka S, Kamiya Y, Jang JC (2011) The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J 65:253–268
Liu XM, Nguyen XC, Kim KE, Han HJ, Yoo J, Lee K, Kim MC, Yun DJ, Chung WS (2013) Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem Biophys Res Commun 430:1054–1059
Monosi B, Wisser RJ, Pennill L, Hulbert SH (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109:1434–1447
Moudouma CF, Riou C, Gloaguen V, Saladin G (2013) Hybrid larch (Larix × eurolepis Henry): a good candidate for cadmium phytoremediation? Environ Sci Pollut Res Int 20:1889–1894
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob-ur-Rahman Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Pomeranz M, Hah C, Lin PC, Kang SG, Finer JJ, Blackshear PJ, Jang JC (2010) The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol 152:151–165
Qiu Q, Ma T, Hu Q, Liu B, Wu Y, Zhou H, Wang Q, Wang J, Liu J (2011) Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol 31:452–461
Sanseverino W, Roma G, De Simone M, Faino L, Melito S, Stupka E, Frusciante L, Ercolano MR (2010) PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res 38:814–821
Sato M, Seki K, Kita K, Moriguchi Y, Yunoki K, Kofujita H, Ohnishi M (2008) Prominent differences in leaf fatty acid composition in the F1 hybrid compared with parent trees Larix gmelinii var. japonica and Larix kaempferi. Biosci Biotechnol Biochem 72:2895–2902
Shu Y, Yu D, Wang D, Guo D, Guo C (2013) Genome-wide survey and expression analysis of the MADS-box gene family in soybean. Mol Biol Rep 40:3901–3911
Song GS, Zhai HL, Peng YG, Zhang L, Wei G, Chen XY, Xiao YG, Wang L, Chen YJ, Wu B, Chen B, Zhang Y, Chen H, Feng XJ, Gong WK, Liu Y, Yin ZJ, Wang F, Liu GZ, Xu HL, Wei XL, Zhao XL, Ouwerkerk PB, Hankemeier T, Reijmers T, van der Heijden R, Lu CM, Wang M, van der Greef J, Zhu Z (2010) Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol Plant 3:1012–1025
Sun JQ, Jiang HL, Xu YX, Li HM, Wu XY, Xie Q, Li CY (2007) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48:1148–1158
Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446:640–645
Torales SL, Rivarola M, Pomponio MF, Gonzalez S, Acuña CV, Fernández P, Lauenstein DL, Verga AR, Hopp HE, Paniego NB, Poltri SN (2013) De novo assembly and characterization of leaf transcriptome for the development of functional molecular markers of the extremophile multipurpose tree species Prosopis alba. BMC Genom 14:705
Trick M, Long Y, Meng J, Bancroft I (2009) Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol J 7:334–346
Wan H, Zhao Z, Malik AA, Qian C, Chen J (2010) Identification and characterization of potential NBS-encoding resistance genes and induction kinetics of a putative candidate gene associated with downy mildew resistance in Cucumis. BMC Plant Biol 10:186
Wan HJ, Yuan W, Bo HL, Shen J, Pang X, Chen JF (2013) Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom 14:109
Wang D, Guo YH, Wu CG, Yang GD, Li YY, Zheng CC (2008) Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom 9:44
Wang Y, Deng D, Bian Y, Lv Y, Xie Q (2010) Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Mol Biol Rep 37:3991–4001
Wang F, Li L, Li H, Liu L, Zhang Y, Gao J, Wang X (2012) Transcriptome analysis of rosette and folding leaves in Chinese cabbage using high-throughput RNA sequencing. Genomics 99:299–307
Watanabe M, Watanabe Y, Kitaoka S, Utsugi H, Kita K, Koike T (2011) Growth and photosynthetic traits of hybrid larch F1 (Larix gmelinii var. japonica × L. kaempferi) under elevated CO2 concentration with low nutrient availability. Tree Physiol 31:965–975
Xu Q, Biswas MK, Lan H, Zeng W, Liu C, Xu J, Deng X (2011) Phylogenetic and evolutionary analysis of NBS-encoding genes in Rutaceae fruit crops. Mol Genet Genomics 285:151–161
Xu DL, Long H, Liang JJ, Zhang J, Chen X, Li JL, Pan ZF, Deng GB, Yu MQ (2012) De novo assembly and characterization of the root transcriptome of Aegilops variabilis during an interaction with the cereal cyst nematode. BMC Genom 13:133
Xue JY, Wang Y, Wu P, Wang Q, Yang LT, Pan XH, Wang B, Chen JQ (2012) A primary survey on bryophyte species reveals two novel classes of nucleotide-binding site (NBS) genes. PLoS ONE 7:e36700
Yang S, Zhang X, Yue JX, Tian D, Chen JQ (2008a) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280:187–198
Yang S, Gu T, Pan C, Feng Z, Ding J, Hang Y, Chen JQ, Tian D (2008b) Genetic variation of NBS-LRR class resistance genes in rice lines. Theor Appl Genet 116:165–177
Zhang Y, Wang L (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5:1
Zhang J, Liang S, Duan J, Wang J, Chen S, Cheng Z, Zhang Q, Liang X, Li Y (2012) De novo assembly and characterisation of the transcriptome during seed development, and generation of genic-SSR markers in Peanut (Arachis hypogaea L.). BMC Genom 13:90
Zhang LF, Li WF, Han SY, Yang WH, Qi LW (2013a) cDNA cloning, genomic organization and expression analysis during somatic embryogenesis of the translationally controlled tumor protein (TCTP) gene from Japanese larch (Larix leptolepis). Gene 529:150–158
Zhang C, Zhang H, Zhao Y, Jiang H, Zhu S, Cheng B, Xiang Y (2013b) Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep 32:1543–1555
Acknowledgments
We thank Dr. Liwang Qi at the Chinese Academy of Forestry, Beijing, China for kindly providing the Larix kaempferi parental lines (Larix13 and Larix82) and their reciprocal hybrids (Larix13 × Larix82-21 and Larix82 × Larix13-6). This work was performed with financial support from the National Natural Science Foundation of China (Nos. 31100234, 31300564 and 31371249) and the Natural Science Foundation of Tianjin (Nos. 14JCQNJC15000, 13JCZDJC29000 and 15JCQNJC15100).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, A., Wang, J., Li, H. et al. Transcriptome profiling and characterization of gene families with zinc finger and nucleotide binding site (NBS) domains in Larix kaempferi . J. Plant Biochem. Biotechnol. 26, 149–159 (2017). https://doi.org/10.1007/s13562-016-0375-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-016-0375-5