Abstract
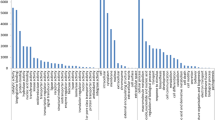
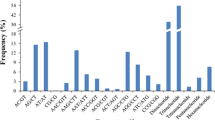
Brassica juncea is an economically important oilseed crop worldwide. It has limited genomic resources at present. We generated 47,962,057 expressed sequence reads which were assembled into 45,280 unigenes. A total of 4108 SSR loci (≥10 bp) were identified in these unigenes. Trinucleotide was the most frequent repeat unit (59.91 %) followed by di- (38.66 %), tetra - (0.71 %), hexa - (0.49 %) and pentanucleotide repeats (0.24 %). Primers were designed for 2863 SSR loci among which 460 were selected for primer synthesis. A total of 339 loci amplified successfully of which 134 (39.5 %) exhibited polymorphism among six B. juncea genotypes with PIC values ranging from 0.18 to 0.81. Further, 25 polymorphic SSRs were used for analysis of genetic variability in 25 genotypes of Brassicas and their wild relatives. Two to five alleles with PIC values 0.22–0.66 were detected at these loci. The dendrogram grouped the genotypes according to their known pedigree/systematic position.




Similar content being viewed by others
References
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Bhardwaj AR, Joshi G, Kukreja B, Malik V, Arora P, Pandey R, Shukla RN, Bankar KG, Agarwal SK et al (2015) Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol 15:9
Bisht NC, Gupta V, Ramchiary N, Sodhi YS, Mukhopadhyay A, Arumugam N, Pental D, Pradhan AK (2009) Fine mapping of loci involved with glucosinolate biosynthesis in oilseed mustard (Brassica juncea) using genomic information from allied species. Theor Appl Genet 118:413–421
Chauhan JS, Singh KH, Singh VV, Satyanshu K (2011) Hundred years of rapeseed-mustard breeding in India: accomplishments and future strategies. Indian J Agric Sci 81:093–1109
Cheung WY, Gugel RK, Landry BS (1998) Identification of RFLP markers linked to the white rust resistance gene (Acr) in mustard (Brassica juncea (L.) Czern. and Coss.). Genome 41:626–628
Dutta S, Kumawat G, Singh BP, Gupta DK, Singh S, Dogra V, Gaikwad K, Sharma TR et al (2011) Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol 11:17
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132
Emrich SJ, Barbazuk WB, Li L, Schnable PS (2007) Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res 17:69–73
Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18:53–63
Getinet A, Sharma SM (eds) (1996) Niger [Guizotia abyssinica (L.f.) Cass.]: promoting the conservation and use of underutilized and neglected crops. International Plant Genetic Resources Institute, Rome
Gómez-Campo C (1999) Taxonomy. In: Biology of Brassica Coenospecies. Elsevier, Amsterdam
Hendre PS, Aggarwal RK (2014) Development of genic and genomic SSR markers of robusta coffee (Coffea canephora Pierre Ex A. Froehner). PLoS One 9(12), e113661
Hopkins CJ, Cogan NOI, Hand M, Jewell E, Kaur J et al (2007) Sixteen new simple sequence repeat markers from Brassica juncea expressed sequences and their cross-species amplification. Mol Ecol Notes 7:697–700
Izzah NK, Lee J, Jayakodi M, Perumal S, Jin M, Beom-Seok P, Ahn K, Yang TJ (2014) Transcriptome sequencing of two parental lines of cabbage (Brassica oleracea L. var. capitata L.) and construction of an EST-based genetic map. BMC Genomics 15:149
Li H, Younas M, Wang X, Li X, Chen L, Zhao B, Chen X, Xu J, Hou F, Hong B, Liu G, Zhao H, Wu X, Du H, Wu J, Liu K (2013) Development of a core set of single-locus SSR markers for allotetraploid rapeseed (Brassica napus L.). Theor Appl Genet 126:937–947
Liu A, Wang J (2006) Genomic evolution of Brassica allopolyploids revealed by ISSR marker. Genet Resour Crop Ev 53:603–611
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15:516–525
Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I et al (2009) Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci 106:12353–12358
Metzgar D, Bytof J, Wills C (2000) Selection against frame shift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80
Mukherjee AK, Mohapatra T, Varshney A, Sharma R, Sharma RP (2001) Molecular mapping of a locus controlling resistance to Albugo candida in Indian mustard. Plant Breed 120:483–487
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Nagaharu U (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci 76:5269–5273
Panjabi-Massand P, Yadava SK, Sharma P, Kaur A, Kumar A, Arumugam N, Sodhi YS, Mukhopadhyay A, Gupta V, Pradhan AK, Pental D (2010) Molecular mapping reveals two independent loci conferring resistance to Albugo candida in the east European germplasm of oilseed mustard Brassica juncea. Theor Appl Genet 121:137–145. doi:10.1007/s00122-010-1297-6
Paritosh K, Yadava SK, Gupta V, Panjabi-Massand P, Sodhi YS, Pradhan AK, Pental D (2013) RNA-seq based SNPs in some agronomically important oleiferous lines of Brassica rapa and their use for genome-wide linkage mapping and specific-region fine mapping. BMC Genomics 14:463
Pavlicek A, Hrda S, Flegr J (1999) Free-Tree–freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol (Praha) 45:97–99
Pradhan AK, Sodhi YS, Mukhopadhyay A, Pental D (1993) Heterosis breeding in Indian mustard (Brassia juncea L. Czern & Coss): analysis of component characters contributing to heterosis for yield. Euphytica 69:219–229
Raman H, Dalton-Morgan J, Diffey S, Raman R, Alamery S, Edwards D, Batley J (2014) SNP markers-based map construction and genome-wide linkage analysis in Brassica napus. Plant Biotechnol J 12:851–860
Ramchiary N, Nguyen VD, Li X, Hong CP, Dhandapani V, Choi SR, Yu G, Piao ZY, Lim YP (2011) Genic microsatellite markers in Brassica rapa: development, characterization, mapping, and their utility in other cultivated and wild Brassica relatives. DNA Res 18:305–320
Roy SW, Penny D, Neafsey DE (2007) Evolutionary conservation of UTR intron boundaries in Cryptococcus. Mol Biol Evol 24:1140–1148
Sabharwal V, Negi MS, Banga SS, Lakshmikumaran M (2004) Mapping of AFLP markers linked to seed coat colour loci in Brassica juncea (L.) Czern. Theor Appl Genet 109:160–166
Sánchez-Yélamo MD (2004) Taxonomic relationships among Erucastrum and Brassica species based on flavonoids compounds. Eucarpia Cruciferae Newsl 25:13–14
Sharma R, Aggarwal RA, Kumar R, Mohapatra T, Sharma RP (2002) Construction of an RAPD linkage map and localization of QTLs for oleic acid level using recombinant inbreds in mustard (Brassica juncea). Genome 45:467–472
Shcheglov AS, Zhulidov PA, Bogdanova EA, Shagin DA (2007) Normalization of cDNA libraries. In: Nucleic acids hybridization. Springer, p 97–124
Singh BK, Thakur AK, Rai PK (2012a) Genetic diversity and relationships in wild species of Brassica and allied genera as revealed by cross-transferable genomic STMS marker assays. Aust J Crop Sci 6:815–821
Singh BK, Thakur AK, Tiwari SK, Siddiqui SA, Singh VV, Rai PK (2012b) Transferability of Brassica-derived microsatellites to related genera and their implications for phylogenetic analysis. Natl Acad Sci Lett 35:37–44
Spect CE, Diederichsen A (2001) Brassica. In: Mansfeld’s encyclopedia of agricultural and horticultural crops. Springer, p 1453–1456
Suwabe K, Iketani H, Nunome T, Kage T, Hirai M (2002) Isolation and characterization of microsatellites in Brassica rapa L. Theor Appl Genet 104:1092–1098
Varshney RK, Graner A, Sorrells ME (2005a) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Varshney RK, Sigmund R, Borner A, Korzun V, Stein N, Sorrells ME, Longridge P, Graner A (2005b) Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci 168:195–202
Victoria FC, da Maia LC, de Oliveira AC (2011) In silico comparative analysis of SSR markers in plants. BMC Plant Biol 11:15
Wang F, Wang XF, Chen X, Xiao Y, Li H, Zhang S, Xu J, Fu J, Huang L, Liu C, Wu J, Liu K (2012) Abundance, marker development and genetic mapping of microsatellites from unigenes in Brassica napus. Mol Breeding 30:731–744
Warwick SI, Black LD (1991) Molecular systematic of Brassica and allied genera (subtribe Brassicinae, Brassiceae)-chloroplast genome and cytodome congruence. Theor Appl Genet 82:81–92
Warwick SI, Black LD (1993) Molecular relationships in subtribe Brassicinae (Cruciferae, tribe Brassiceae). Can J Bot 71:906–918
Weber JL (1990) Informativeness of human (dC - dA)n (dG - dT)n polymorphisms. Genomics 7:524–530
Wei W, Qi X, Wang L, Zhang Y, Hua W, Li D, Haixia L, Zhang X (2011) Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics 12:451
Yu JK, La Rota M, Kantety RV, Sorrells ME (2004a) EST derived SSR markers for comparative mapping in wheat and rice. Mol Gen Genet 271:742–751
Yu JK, Dake TM, Singh S, Benscher D, Li W, Gill B, Sorrells ME (2004b) Development and mapping of EST-derived simple sequence repeat (SSR) markers for hexaploid wheat. Genome 47:805–818
Zhang H, Wei L, Miao H, Zhang T, Wang C (2012) Development and validation of genic-SSR markers in sesame by RNA-seq. BMC Genomics 13:316
Acknowledgments
We sincerely acknowledge Director, ICAR-Directorate of Rapeseed-Mustard Research, Bharatpur-321303, Rajasthan, India, for providing financial support and the facilities to carry out this research work. We also gratefully acknowledge Director, ICAR - Indian Agricultural Statistics Research Institute, New Delhi - 110 012, India, for providing computational facilities of Centre for Agricultural Bioinformatics, and Dr. S.R. Bhatt (Principal Scientist), ICAR- National Research Centre on Plant Biotechnology, New Delhi – 110012, India, for providing seed materials of wild species used in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interest.
Electronic supplementary material
Supplementary Information Available
Supplementary information is included with this article.
Supplementary Table 1
Brassicas and wild relative species used for the validation of genic-SSRs (DOC 59 kb)
Supplementary Table 2
Particulars of 2863 genic-SSR loci (≥10 bp) in Brassica juncea for which primers were designed (XLS 629 kb)
Supplementary Table 3
Particulars of 339 validated genic-SSRs of Brassica juncea and predicted function of their genes (XLS 116 kb)
Supplementary Table 4
Particulars of 134 genic-SSR loci showing polymorphism among six Brassica juncea genotypes (XLS 37 kb)
Rights and permissions
About this article
Cite this article
Singh, B.K., Mishra, D.C., Yadav, S. et al. Identification, characterization, validation and cross-species amplification of genic-SSRs in Indian Mustard (Brassica juncea). J. Plant Biochem. Biotechnol. 25, 410–420 (2016). https://doi.org/10.1007/s13562-016-0353-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-016-0353-y