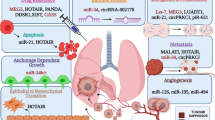
Abstract
Lung cancer, being the most widespread and lethal form of cancer globally, has a high incidence and mortality rate primarily attributed to challenges associated with early detection, extensive metastasis, and frequent recurrence. In the context of lung cancer development, noncoding RNA molecules have a crucial role in governing gene expression and protein synthesis. Specifically, tRNA-derived fragments (tRFs), a subset of noncoding RNAs, exert significant biological influences on cancer progression, encompassing transcription and translation processes as well as epigenetic regulation. This article primarily examines the mechanisms by which tRFs modulate gene expression and contribute to tumorigenesis in lung cancer. Furthermore, we provide a comprehensive overview of the current bioinformatics analysis of tRFs in lung cancer, with the objective of offering a systematic and efficient approach for studying the expression profiling, functional enrichment, and molecular mechanisms of tRFs in this disease. Finally, we discuss the clinical significance and potential avenues for future research on tRFs in lung cancer. This paper presents a comprehensive systematic review of the existing research findings on tRFs in lung cancer, aiming to offer improved biomarkers and drug targets for clinical management of lung cancer.




Similar content being viewed by others
Data Availability
Not applicable.
Code Availability
Not applicable.
Abbreviations
- ψ:
-
pseudouracil
- 3’-UTR:
-
3’-untranslated region
- AGO:
-
Argonaute
- ALKBH3:
-
AlkB homologue 3
- ANG:
-
Angiogenin
- AKT:
-
Protein kinase B
- AURKA:
-
Aurora kinase A
- BCL2L11:
-
B-cell lymphoma 2-like 11
- CELF6:
-
CUGBP Elav-Like Family Member 6
- ceRNA:
-
Competitive endogenous RNA
- circRNAs:
-
Circular RNAs
- D:
-
Dihydrouracil
- EGFR-TKIs:
-
Epidermal growth factor receptor tyrosine kinase inhib-itors
- ELK4:
-
ETS-like transcription factor 4
- EMT:
-
Epithelial-mesenchymal transition
- GADD45G:
-
DNA damage 45G
- GEPIA:
-
Gene Expression Profiling Interactive Analysis
- GEO:
-
Gene Expression Omnibus
- Gm:
-
2’-O-methylguanosine
- GO:
-
Gene Ontology
- GSEA:
-
Gene set enrichment analysis
- H3K27ac:
-
H3-lysine-27-acetylation
- H3K4me1:
-
H3-lysine 4-monomethylation
- HSPB1:
-
Small heat shock protein-1
- i6A:
-
N6-isopentenyl adenosine
- IGF1R:
-
Insulin-like growth factor 1 receptor
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LARS1:
-
Leucine-tRNA synthetase
- lncRNAs:
-
Long noncoding RNAs
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
lung squamous cell carcinoma
- MED29:
-
Mediator complex subunit 29
- miRNAs:
-
MicroRNAs
- Mthfd1l:
-
Methylenetetra-hydrofolate dehydrogenase 1 like
- ncRNAs:
-
Noncoding RNAs
- NSun2:
-
NOP2/Sun RNA methyltransferase 2
- NSCLC:
-
Non-small cell lung cancer
- Pafah1b1:
-
Platelet-activating factor acetylhydrolase 1 beta 1
- PIWIL2:
-
Piwi-like protein 2
- RBPs:
-
RNA-binding proteins
- RIP:
-
RNA immunoprecipitation
- RISC:
-
RNA-induced silencing complex
- RNP:
-
Ribonucleoprotein
- RUNX1:
-
Runt-related transcription factor 1
- SCLC:
-
Small cell lung cancer
- SPRY4:
-
Sprouty 4
- TCGA:
-
The Cancer Genome Atlas
- TEM:
-
Transmission electron microscopy
- TFs:
-
Transcription factors
- tiRNAs:
-
tRNA semimolecules
- tRFs:
-
tRNA-derived fragments
- TRMT2A:
-
tRNA methyltransferase 2 homologue A
- YTHDF1:
-
YTH N6-methyladenosine RNA binding protein 1
References
R.L. Siegel, K.D. Miller, H.E. Fuchs, A. Jemal, Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33 (2022)
N. Duma, R. Santana-Davila, J.R. Molina, Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 94, 1623–1640 (2019)
I.R. Konig, O. Fuchs, G. Hansen, E. von Mutius, M.V. Kopp, What is precision medicine? Eur. Respir J. 50(2017)
B.W. Carter, M. Altan, G.S. Shroff, M.T. Truong, I. Vlahos, Post-chemotherapy and targeted therapy imaging of the chest in lung cancer. Clin. Radiol. 77, e1–e10 (2022)
Y. Chen, Z. Chen, R. Chen, C. Fang, C. Zhang, M. Ji, X. Yang, Immunotherapy-based combination strategies for treatment of EGFR-TKI-resistant non-small-cell lung cancer. Future Oncol. 18, 1757–1775 (2022)
M. Haider, A. Elsherbeny, V. Pittala, V. Consoli, M.A. Alghamdi, Z. Hussain, G. Khoder, K. Greish, Nanomedicine strategies for management of Drug Resistance in Lung Cancer. Int. J. Mol. Sci. 23(2022)
K. O’Leary, A. Shia, P. Schmid, Epigenetic regulation of EMT in Non-Small Cell Lung Cancer. Curr. Cancer Drug Targets. 18, 89–96 (2018)
S. Panni, R.C. Lovering, P. Porras, S. Orchard, Non-coding RNA regulatory networks. Biochim. Biophys. Acta Gene Regul. Mech. 1863, 194417 (2020)
Q. Chen, X. Zhang, J. Shi, M. Yan, T. Zhou, Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem. Sci. 46, 790–804 (2021)
Y. Zhang, X. Zhang, J. Shi, F. Tuorto, X. Li, Y. Liu, R. Liebers, L. Zhang, Y. Qu, J. Qian, M. Pahima, Y. Liu, M. Yan, Z. Cao, X. Lei, Y. Cao, H. Peng, S. Liu, Y. Wang, H. Zheng, R. Woolsey, D. Quilici, Q. Zhai, L. Li, T. Zhou, W. Yan, F. Lyko, Y. Zhang, Q. Zhou, E. Duan, Q. Chen, Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell. Biol. 20, 535–540 (2018)
J. Park, S.H. Ahn, M.G. Shin, H.K. Kim, S. Chang, tRNA-Derived small RNAs: novel epigenetic regulators. Cancers (Basel) 12(2020)
A.N. Shaukat, E.G. Kaliatsi, V. Stamatopoulou, C. Stathopoulos, Mitochondrial tRNA-Derived fragments and their contribution to Gene expression regulation. Front. Physiol. 12, 729452 (2021)
J. Shi, Y. Zhang, T. Zhou, Q. Chen, tsRNAs: the Swiss Army Knife for Translational Regulation. Trends Biochem. Sci. 44, 185–189 (2019)
Z. Su, I. Monshaugen, B. Wilson, F. Wang, A. Klungland, R. Ougland, A. Dutta, TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat. Commun. 13, 2165 (2022)
J.T. Wen, Z.H. Huang, Q.H. Li, X. Chen, H.L. Qin, Y. Zhao, Research progress on the tsRNA classification, function, and application in gynecological malignant tumors. Cell. Death Discovery 7(2021)
B. Chen, S. Liu, H. Wang, G. Li, X. Lu, H. Xu, Differential Expression Profiles and Function Prediction of Transfer RNA-Derived Fragments in High-Grade Serous Ovarian Cancer. Biomed Res Int 2021, 5594081 (2021)
W. Xu, M. Yu, Y. Wu, Y. Jie, X. Li, X. Zeng, F. Yang, Y. Chong, Plasma-derived exosomal SncRNA as a Promising Diagnostic Biomarker for early detection of HBV-Related Acute-on-chronic liver failure. Front. Cell. Infect. Microbiol. 12, 923300 (2022)
X. Liu, Y.Z. Wen, Z.L. Huang, X. Shen, J.H. Wang, Y.H. Luo, W.X. Chen, Z.R. Lun, H.B. Li, L.H. Qu, H. Shan, L.L. Zheng, SARS-CoV-2 causes a significant stress response mediated by small RNAs in the blood of COVID-19 patients. Mol. Ther. Nucleic Acids. 27, 751–762 (2022)
W. Liu, Y. Liu, Z. Pan, X. Zhang, Y. Qin, X. Chen, M. Li, X. Chen, Q. Zheng, X. Liu, D. Li, Systematic analysis of tRNA-Derived small RNAs discloses new therapeutic targets of caloric restriction in myocardial ischemic rats. Front. Cell. Dev. Biol. 8, 568116 (2020)
Z. Ma, J. Zhou, Y. Shao, F.A. Jafari, P. Qi, Y. Li, Biochemical properties and progress in cancers of tRNA-derived fragments. J. Cell. Biochem. 121, 2058–2063 (2020)
O.A. Esakova, T.L. Grove, N.H. Yennawar, A.J. Arcinas, B. Wang, C. Krebs, S.C. Almo, S.J. Booker, Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB. Nature. 597, 566–570 (2021)
S. Rashad, K. Niizuma, T. Tominaga, tRNA cleavage: a new insight. Neural Regen Res. 15, 47–52 (2020)
H. Huang, H. Li, R. Pan, S. Wang, X. Liu, tRNA modifications and their potential roles in pancreatic cancer. Arch. Biochem. Biophys. 714, 109083 (2021)
M. Kazimierczyk, M. Wojnicka, E. Biala, P. Zydowicz-Machtel, B. Imiolczyk, T. Ostrowski, A. Kurzynska-Kokorniak, J. Wrzesinski, Characteristics of transfer RNA-Derived fragments expressed during human renal cell development: the role of Dicer in tRF Biogenesis. Int. J. Mol. Sci. 23(2022)
C.E. Monaghan, S.I. Adamson, M. Kapur, J.H. Chuang, S.L. Ackerman, The Clp1 R140H mutation alters tRNA metabolism and mRNA 3’ processing in mouse models of pontocerebellar hypoplasia. Proc. Natl. Acad. Sci. U. S. A. 118(2021)
Z.X. Huang, J. Li, Q.P. Xiong, H. Li, E.D. Wang, R.J. Liu, Position 34 of tRNA is a discriminative element for m5C38 modification by human DNMT2. Nucleic Acids Res. 49, 13045–13061 (2021)
S. Rashad, X. Han, K. Sato, E. Mishima, T. Abe, T. Tominaga, K. Niizuma, The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 17, 1092–1103 (2020)
Z. Chen, M. Qi, B. Shen, G. Luo, Y. Wu, J. Li, Z. Lu, Z. Zheng, Q. Dai, H. Wang, Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 47, 2533–2545 (2019)
Y. Wu, X. Yang, G. Jiang, H. Zhang, L. Ge, F. Chen, J. Li, H. Liu, H. Wang, 5’-tRF-GlyGCC: a tRNA-derived small RNA as a novel biomarker for colorectal cancer diagnosis. Genome Med. 13, 20 (2021)
F. Pichot, M.C. Hogg, V. Marchand, V. Bourguignon, E. Jirstrom, C. Farrell, H.A. Gibriel, J.H.M. Prehn, Y. Motorin, M. Helm, Quantification of substoichiometric modification reveals global tsRNA hypomodification, preferences for angiogenin-mediated tRNA cleavage, and idiosyncratic epitranscriptomes of human neuronal cell-lines. Comput. Struct. Biotechnol. J. 21, 401–417 (2023)
K.Y. Cao, Y. Pan, T.M. Yan, P. Tao, Y. Xiao, Z.H. Jiang, Antitumor Activities of tRNA-Derived Fragments and tRNA Halves from Non-pathogenic Escherichia coli Strains on Colorectal Cancer and Their Structure-Activity Relationship. mSystems 7, e0016422 (2022)
W. Wu, I. Lee, H. Spratt, X. Fang, X. Bao, tRNA-Derived fragments in Alzheimer’s Disease: implications for New Disease biomarkers and neuropathological mechanisms. J. Alzheimers Dis. 79, 793–806 (2021)
F. Ren, K.Y. Cao, R.Z. Gong, M.L. Yu, P. Tao, Y. Xiao, Z.H. Jiang, The role of post-transcriptional modification on a new tRNA(ile(GAU)) identified from Ganoderma lucidum in its fragments’ cytotoxicity on cancer cells. Int. J. Biol. Macromol. 229, 885–895 (2023)
M. Pereira, D.R. Ribeiro, M.M. Pinheiro, M. Ferreira, S. Kellner, A.R. Soares, M(5)U54 tRNA hypomodification by lack of TRMT2A drives the generation of tRNA-Derived small RNAs. Int. J. Mol. Sci. 22(2021)
A. Molla-Herman, M.T. Angelova, M. Ginestet, C. Carre, C. Antoniewski, J.R. Huynh, tRNA fragments populations analysis in mutants affecting tRNAs Processing and tRNA methylation. Front. Genet. 11, 518949 (2020)
Z. Sun, J. Tan, M. Zhao, Q. Peng, M. Zhou, S. Zuo, F. Wu, X. Li, Y. Dong, M. Xie, Y. Yang, J. Zhou, X. Liu, Q. He, Z. He, X. Yu, Q. He, Integrated genomic analysis reveals regulatory pathways and dynamic landscapes of the tRNA transcriptome. Sci. Rep. 11, 5226 (2021)
A. Liaqat, C. Stiller, M. Michel, M.V. Sednev, C. Hobartner, N(6) -Isopentenyladenosine in RNA determines the cleavage site of endonuclease deoxyribozymes. Angew Chem. Int. Ed. Engl. 59, 18627–18631 (2020)
Z. Liu, H.K. Kim, J. Xu, Y. Jing, M.A. Kay, The 3’tsRNAs are aminoacylated: Implications for their biogenesis. PLoS Genet. 17, e1009675 (2021)
U. Testa, E. Pelosi, G. Castelli, Molecular charcterization of lung adenocarcinoma combining whole exome sequencing, copy number analysis and gene expression profiling. Expert Rev. Mol. Diagn. 22, 77–100 (2022)
F. Santos, A.M. Capela, F. Mateus, S. Nobrega-Pereira, Bernardes de Jesus, non-coding antisense transcripts: fine regulation of gene expression in cancer. Comput. Struct. Biotechnol. J. 20, 5652–5660 (2022)
S. Roy, N. Ganguly, S. Banerjee, Exploring clinical implications and role of non-coding RNAs in lung carcinogenesis. Mol. Biol. Rep. 49, 6871–6883 (2022)
A.S. Doghish, A. Ismail, M.A. Elrebehy, A.M.M. Elbadry, H.H. Mahmoud, S.M. Farouk, G.A.A. Serea, R.A.A. Elghany, K.K. El-Halwany, A.O. Alsawah, H.I. Dewidar, H.A. El-Mahdy, A study of miRNAs as cornerstone in lung cancer pathogenesis and therapeutic resistance: a focus on signaling pathways interplay. Pathol. Res. Pract. 237(2022)
X. Zhang, X. Wang, B. Chai, Z. Wu, X. Liu, H. Zou, Z. Hua, Z. Ma, W. Wang, Downregulated miR-18a and miR-92a synergistically suppress non-small cell lung cancer via targeting sprouty 4. Bioengineered. 13, 11281–11295 (2022)
J. Hua, X. Wang, L. Ma, J. Li, G. Cao, S. Zhang, W. Lin, CircVAPA promotes small cell lung cancer progression by modulating the mir-377-3p and miR-494-3p/IGF1R/AKT axis. Mol. Cancer. 21, 123 (2022)
M. Yu, B. Lu, J. Zhang, J. Ding, P. Liu, Y. Lu, tRNA-derived RNA fragments in cancer: current status and future perspectives. J. Hematol. Oncol. 13, 121 (2020)
L. Ye, F. Wang, J. Wang, H. Wu, H. Yang, Z. Yang, H. Huang, Role and mechanism of miR-211 in human cancer. J. Cancer. 13, 2933–2944 (2022)
J. Shen, Y. Wu, W. Ruan, F. Zhu, S. Duan, miR-1908 Dysregulation in Human Cancers. Front. Oncol. 12, 857743 (2022)
T.T.P. Nguyen, K.H. Suman, T.B. Nguyen, H.T. Nguyen, D.N. Do, The Role of miR-29s in Human Cancers-An Update. Biomedicines 10(2022)
W. Meng, Y. Li, B. Chai, X. Liu, Z. Ma, miR-199a: a tumor suppressor with noncoding RNA network and therapeutic candidate in Lung Cancer. Int. J. Mol. Sci. 23(2022)
L. Zhang, Z. Yang, A. Ma, Y. Qu, S. Xia, D. Xu, C. Ge, B. Qiu, Q. Xia, J. Li, Y. Liu, Growth arrest and DNA damage 45G down-regulation contributes to Janus kinase/signal transducer and activator of transcription 3 activation and cellular senescence evasion in hepatocellular carcinoma. Hepatology. 59, 178–189 (2014)
F. Hu, Y. Niu, X. Mao, J. Cui, X. Wu, C.B. 2 Simone nd, H.S. Kang, W. Qin, L. Jiang, tsRNA-5001a promotes proliferation of lung adenocarcinoma cells and is associated with postoperative recurrence in lung adenocarcinoma patients. Transl Lung Cancer Res. 10, 3957–3972 (2021)
H. Fan, H. Liu, Y. Lv, Y. Song, AS-tDR-007872: A Novel tRNA-Derived Small RNA Acts an Important Role in Non-Small-Cell Lung Cancer. Comput. Math. Methods Med. 2022, 3475955 (2022)
X. Li, D. Zhang, B. Li, B. Zou, S. Wang, B. Fan, W. Li, J. Yu, L. Wang, Clinical implications of germline BCL2L11 deletion polymorphism in pretreated advanced NSCLC patients with osimertinib therapy. Lung Cancer. 151, 39–43 (2021)
H. Ma, G. Liu, B. Yu, J. Wang, Y. Qi, Y. Kou, Y. Hu, S. Wang, F. Wang, D. Chen, RNA-binding protein CELF6 modulates transcription and splicing levels of genes associated with tumorigenesis in lung cancer A549 cells. PeerJ 10, e13800 (2022)
L. Ma, X. Xue, X. Zhang, K. Yu, X. Xu, X. Tian, Y. Miao, F. Meng, X. Liu, S. Guo, S. Qiu, Y. Wang, J. Cui, W. Guo, Y. Li, J. Xia, Y. Yu, J. Wang, The essential roles of m(6)a RNA modification to stimulate ENO1-dependent glycolysis and tumorigenesis in lung adenocarcinoma. J. Exp. Clin. Cancer Res. 41, 36 (2022)
J. Wang, Z. Wang, W. Lin, Q. Han, H. Yan, W. Yao, R. Dong, D. Jia, K. Dong, K. Li, LINC01296 promotes neuroblastoma tumorigenesis via the NCL-SOX11 regulatory complex. Mol. Ther. Oncolytics. 24, 834–848 (2022)
N.C. Zetouni, C.M. Sergi, in Metastasis, ed. by C.M. Sergi (Brisbane (AU), 2022)
X. Liu, W. Mei, V. Padmanaban, H. Alwaseem, H. Molina, M.C. Passarelli, B. Tavora, S.F. Tavazoie, A pro-metastatic tRNA fragment drives Nucleolin oligomerization and stabilization of its bound metabolic mRNAs. Mol. Cell 82, 2604–2617 e2608 (2022)
H. Shaath, R. Vishnubalaji, R. Elango, A. Kardousha, Z. Islam, R. Qureshi, T. Alam, P.R. Kolatkar, N.M. Alajez, Long non-coding RNA and RNA-binding protein interactions in cancer: experimental and machine learning approaches. Semin. Cancer Biol. 86, 325–345 (2022)
S.A. Lachke, RNA-binding proteins and post-transcriptional regulation in lens biology and cataract: mediating spatiotemporal expression of key factors that control the cell cycle, transcription, cytoskeleton and transparency. Exp. Eye Res. 214, 108889 (2022)
W. Yang, K. Gao, Y. Qian, Y. Huang, Q. Xiang, C. Chen, Q. Chen, Y. Wang, F. Fang, Q. He, S. Chen, J. Xiong, Y. Chen, N. Xie, D. Zheng, R. Zhai, A novel tRNA-derived fragment AS-tDR-007333 promotes the malignancy of NSCLC via the HSPB1/MED29 and ELK4/MED29 axes. J. Hematol. Oncol. 15, 53 (2022)
W. Zhang, X. Ruan, Y. Li, J. Zhi, L. Hu, X. Hou, X. Shi, X. Wang, J. Wang, W. Ma, P. Gu, X. Zheng, M. Gao, KDM1A promotes thyroid cancer progression and maintains stemness through the Wnt/beta-catenin signaling pathway. Theranostics. 12, 1500–1517 (2022)
A.D. Durbin, T. Wang, V.K. Wimalasena, M.W. Zimmerman, D. Li, N.V. Dharia, L. Mariani, N.A.M. Shendy, S. Nance, A.G. Patel, Y. Shao, M. Mundada, L. Maxham, P.M.C. Park, L.H. Sigua, K. Morita, A.S. Conway, A.L. Robichaud, A.R. Perez-Atayde, M.J. Bikowitz, T.R. Quinn, O. Wiest, J. Easton, E. Schonbrunn, M.L. Bulyk, B.J. Abraham, K. Stegmaier, A.T. Look, J., Qi, EP300 Selectively Controls the Enhancer Landscape of MYCN-Amplified Neuroblastoma. Cancer Discov. 12, 730–751 (2022)
X. Gu, Y. Zhang, X. Qin, S. Ma, Y. Huang, S. Ju, Transfer RNA-derived small RNA: an emerging small non-coding RNA with key roles in cancer. Exp. Hematol. Oncol. 11, 35 (2022)
W.J. de Jonge, H.P. Patel, J.V.W. Meeussen, T.L. Lenstra, Following the tracks: how transcription factor binding dynamics control transcription. Biophys. J. 121, 1583–1592 (2022)
R.W.J. Wong, J.Z.L. Ong, M.S. Theardy, T. Sanda, IRF4 as an oncogenic master transcription factor. Cancers (Basel) 14(2022)
P. Debnath, R.S. Huirem, P. Dutta, S. Palchaudhuri, Epithelial-mesenchymal transition and its transcription factors. Biosci. Rep. 42(2022)
L. Shen, Z. Tan, M. Gan, Q. Li, L. Chen, L. Niu, D. Jiang, Y. Zhao, J. Wang, X. Li, S. Zhang, L. Zhu, tRNA-Derived Small Non-Coding RNAs as Novel Epigenetic Molecules Regulating Adipogenesis. Biomolecules 9(2019)
N.H. Farina, S. Scalia, C.E. Adams, D. Hong, A.J. Fritz, T.L. Messier, V. Balatti, D. Veneziano, J.B. Lian, C.M. Croce, G.S. Stein, J.L. Stein, Identification of tRNA-derived small RNA (tsRNA) responsive to the tumor suppressor, RUNX1, in breast cancer. J. Cell. Physiol. 235, 5318–5327 (2020)
L. Zheng, H. Xu, Y. Di, L. Chen, J. Liu, L. Kang, L. Gao, ELK4 promotes the development of gastric cancer by inducing M2 polarization of macrophages through regulation of the KDM5A-PJA2-KSR1 axis. J. Transl Med. 19, 342 (2021)
M. Chen, Y. Liu, Y. Yang, Y. Qiu, Z. Wang, X. Li, W. Zhang, Emerging roles of activating transcription factor (ATF) family members in tumourigenesis and immunity: implications in cancer immunotherapy. Genes Dis. 9, 981–999 (2022)
X. Hu, J. Miao, M. Zhang, X. Wang, Z. Wang, J. Han, D. Tong, C. Huang, miRNA-103a-3p promotes human gastric Cancer cell proliferation by Targeting and suppressing ATF7 in vitro. Mol. Cells. 41, 390–400 (2018)
V. Balatti, G. Nigita, D. Veneziano, A. Drusco, G.S. Stein, T.L. Messier, N.H. Farina, J.B. Lian, L. Tomasello, C.G. Liu, A. Palamarchuk, J.R. Hart, C. Bell, M. Carosi, E. Pescarmona, L. Perracchio, M. Diodoro, A. Russo, A. Antenucci, P. Visca, A. Ciardi, C.C. Harris, P.K. Vogt, Y. Pekarsky, C.M. Croce, tsRNA signatures in cancer. Proc. Natl. Acad. Sci. U. S. A. 114, 8071–8076 (2017)
X. Sun, J. Yang, M. Yu, D. Yao, L. Zhou, X. Li, Q. Qiu, W. Lin, B. Lu, E. Chen, P. Wang, W. Chen, S. Tao, H. Xu, A. Williams, Y. Liu, X. Pan, A.W. Jr. Cowley, W. Lu, M. Liang, P. Liu, Y. Lu, Global identification and characterization of tRNA-derived RNA fragment landscapes across human cancers. NAR Cancer. 2, zcaa031 (2020)
Y. Shao, Q. Sun, X. Liu, P. Wang, R. Wu, Z. Ma, tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem. Biol. Drug Des. 90, 730–738 (2017)
N.J. Taylor, J.T. Bensen, C. Poole, M.A. Troester, M.D. Gammon, J. Luo, R.C. Millikan, A.F. Olshan, Genetic variation in cell cycle regulatory gene AURKA and association with intrinsic breast cancer subtype. Mol. Carcinog. 54, 1668–1677 (2015)
M. Zhang, C. Huo, Y. Jiang, J. Liu, Y. Yang, Y. Yin, Y. Qu, AURKA and FAM83A are prognostic biomarkers and correlated with tumor-infiltrating lymphocytes in smoking related Lung Adenocarcinoma. J. Cancer. 12, 1742–1754 (2021)
X. Gong, J. Du, S.H. Parsons, F.F. Merzoug, Y. Webster, P.W. Iversen, L.C. Chio, R.D. Van Horn, X. Lin, W. Blosser, B. Han, S. Jin, S. Yao, H. Bian, C. Ficklin, L. Fan, A. Kapoor, S. Antonysamy, A.M. Mc Nulty, K. Froning, D. Manglicmot, A. Pustilnik, K. Weichert, S.R. Wasserman, M. Dowless, C. Marugan, C. Baquero, M.J. Lallena, S.W. Eastman, Y.H. Hui, M.Z. Dieter, T. Doman, S. Chu, H.R. Qian, X.S. Ye, D.A. Barda, G.D. Plowman, C. Reinhard, R.M. Campbell, J.R. Henry, Buchanan, Aurora a kinase inhibition is Synthetic Lethal with loss of the RB1 tumor suppressor gene. Cancer Discov. 9, 248–263 (2019)
W. Du, T.L. Frankel, M. Green, W. Zou, IFNgamma signaling integrity in colorectal cancer immunity and immunotherapy. Cell. Mol. Immunol. 19, 23–32 (2022)
B. Taciak, I. Pruszynska, L. Kiraga, M. Bialasek, M. Krol, Wnt signaling pathway in development and cancer. J. Physiol. Pharmacol. 69(2018)
Y. Zheng, L. Wang, L. Yin, Z. Yao, R. Tong, J. Xue, Y. Lu, Lung Cancer Stem cell markers as therapeutic targets: an update on Signaling Pathways and Therapies. Front. Oncol. 12, 873994 (2022)
J.Y. Zheng, C. Li, Z.N. Zhu, F.M. Yang, X.M. Wang, P. Jiang, F. Yan, A 5’-tRNA derived Fragment named tiRNA-Val-CAC-001 works as a suppressor in gastric Cancer. Cancer Manag. Res. 14, 2323–2337 (2022)
D. Mo, P. Jiang, Y. Yang, X. Mao, X. Tan, X. Tang, D. Wei, B. Li, X. Wang, L. Tang, F. Yan, A tRNA fragment, 5’-tiRNA(val), suppresses the Wnt/beta-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett. 457, 60–73 (2019)
L. Zhu, Z. Li, X. Yu, Y. Ruan, Y. Shen, Y. Shao, X. Zhang, G. Ye, J. Guo, The tRNA-derived fragment 5026a inhibits the proliferation of gastric cancer cells by regulating the PTEN/PI3K/AKT signaling pathway. Stem Cell. Res. Ther. 12, 418 (2021)
W. Xu, B. Zhou, J. Wang, L. Tang, Q. Hu, J. Wang, H. Chen, J. Zheng, F. Yan, H. Chen, tRNA-Derived fragment tRF-Glu-TTC-027 regulates the progression of gastric carcinoma via MAPK signaling pathway. Front. Oncol. 11, 733763 (2021)
B. Huang, H. Yang, X. Cheng, D. Wang, S. Fu, W. Shen, Q. Zhang, L. Zhang, Z. Xue, Y. Li, Y. Da, Q. Yang, Z. Li, L. Liu, L. Qiao, Y. Kong, Z. Yao, P. Zhao, M. Li, R. Zhang, tRF/miR-1280 suppresses stem cell-like cells and metastasis in Colorectal Cancer. Cancer Res. 77, 3194–3206 (2017)
E.W. Tao, H.L. Wang, W.Y. Cheng, Q.Q. Liu, Y.X. Chen, Q.Y. Gao, A specific tRNA half, 5’tiRNA-His-GTG, responds to hypoxia via the HIF1alpha/ANG axis and promotes colorectal cancer progression by regulating LATS2. J. Exp. Clin. Cancer Res. 40, 67 (2021)
D. Mo, F. He, J. Zheng, H. Chen, L. Tang, F. Yan, tRNA-Derived fragment tRF-17-79MP9PP attenuates Cell Invasion and Migration via THBS1/TGF-beta1/Smad3 Axis in breast Cancer. Front. Oncol. 11, 656078 (2021)
Y.Y. Xie, L.P. Yao, X.C. Yu, Y. Ruan, Z. Li, J.M. Guo, Action mechanisms and research methods of tRNA-derived small RNAs. Signal Transduct. Target. Therapy 5(2020)
V. Pliatsika, P. Loher, R. Magee, A.G. Telonis, E. Londin, M. Shigematsu, Y. Kirino, I. Rigoutsos, MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all the Cancer Genome Atlas projects. Nucleic Acids Res. 46, D152–D159 (2018)
J. Ma, F. Liu, Study of tRNA-Derived Fragment tRF-20-S998LO9D in Pan-Cancer. Dis. Markers 2022, 8799319 (2022)
Z. Gao, M. Jijiwa, M. Nasu, H. Borgard, T. Gong, J. Xu, S. Chen, Y. Fu, Y. Chen, X. Hu, G. Huang, Y. Deng, Comprehensive landscape of tRNA-derived fragments in lung cancer. Mol. Ther. Oncolytics. 26, 207–225 (2022)
J. Zhang, L. Li, L. Luo, X. Yang, J. Zhang, Y. Xie, R. Liang, W. Wang, S. Lu, Screening and potential role of tRFs and tiRNAs derived from tRNAs in the carcinogenesis and development of lung adenocarcinoma. Oncol. Lett. 22, 506 (2021)
J. Wang, X. Liu, W. Cui, Q. Xie, W. Peng, H. Zhang, Y. Gao, C. Zhang, C. Duan, Plasma tRNA-derived small RNAs signature as a predictive and prognostic biomarker in lung adenocarcinoma. Cancer Cell. Int. 22, 59 (2022)
S.R. Yang, A.M. Schultheis, H. Yu, D. Mandelker, M. Ladanyi, R. Buttner, Precision medicine in non-small cell lung cancer: current applications and future directions. Semin. Cancer Biol. 84, 184–198 (2022)
K. Christofyllakis, A.R. Monteiro, O. Cetin, I.A. Kos, A. Greystoke, A. Luciani, Biomarker guided treatment in oncogene-driven advanced non-small cell lung cancer in older adults: a Young International Society of Geriatric Oncology report. J. Geriatr. Oncol. 13, 1071–1083 (2022)
C. Zhong, Z. Xie, L.H. Zeng, C. Yuan, S. Duan, MIR4435-2HG is a potential Pan-Cancer Biomarker for diagnosis and prognosis. Front. Immunol. 13, 855078 (2022)
C. Zhong, Z. Xie, J. Shen, Y. Jia, S. Duan, LINC00665: An Emerging Biomarker for Cancer Diagnostics and Therapeutics. Cells 11, (2022)
Y. Jia, W. Tan, Y. Zhou, Transfer RNA-derived small RNAs: potential applications as novel biomarkers for disease diagnosis and prognosis. Ann. Transl Med. 8, 1092 (2020)
T. Zong, Y. Yang, H. Zhao, L. Li, M. Liu, X. Fu, G. Tang, H. Zhou, L.H.H. Aung, P. Li, J. Wang, Z. Wang, T. Yu, tsRNAs: Novel small molecules from cell function and regulatory mechanism to therapeutic targets. Cell Prolif. 54, e12977 (2021)
S.U. Umu, H. Langseth, V. Zuber, A. Helland, R. Lyle, T.B. Rounge, Serum RNAs can predict lung cancer up to 10 years prior to diagnosis. Elife 11(2022)
W. Gu, J. Shi, H. Liu, X. Zhang, J.J. Zhou, M. Li, D. Zhou, R. Li, J. Lv, G. Wen, S. Zhu, T. Qi, W. Li, X. Wang, Z. Wang, H. Zhu, C. Zhou, K.S. Knox, T. Wang, Q. Chen, Z. Qian, T. Zhou, Peripheral blood non-canonical small non-coding RNAs as novel biomarkers in lung cancer. Mol. Cancer. 19, 159 (2020)
J. You, G. Yang, Y. Wu, X. Lu, S. Huang, Q. Chen, C. Huang, F. Chen, X. Xu, L. Chen, Plasma tRF-1:29-Pro-AGG-1-M6 and tRF-55:76-Tyr-GTA-1-M2 as novel diagnostic biomarkers for lung adenocarcinoma. Front. Oncol. 12, 991451 (2022)
J. Li, C. Cao, L. Fang, W. Yu, Serum transfer RNA-derived fragment tRF-31-79MP9P9NH57SD acts as a novel diagnostic biomarker for non-small cell lung cancer. J. Clin. Lab. Anal. 36(2022)
R. Kalluri, V.S. LeBleu, The biology, function, and biomedical applications of exosomes. Science 367(2020)
H. Xie, J. Yao, Y. Wang, B. Ni, Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 29, 1257–1271 (2022)
S. Liu, W. Wang, Y. Ning, H. Zheng, Y. Zhan, H. Wang, Y. Yang, J. Luo, Q. Wen, H. Zang, J. Peng, J. Ma, S. Fan, Exosome-mediated mir-7-5p delivery enhances the anticancer effect of Everolimus via blocking MNK/eIF4E axis in non-small cell lung cancer. Cell. Death Dis. 13, 129 (2022)
M.Y. Li, L.Z. Liu, M. Dong, Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol. Cancer. 20, 22 (2021)
B. Zheng, X. Song, L. Wang, Y. Zhang, Y. Tang, S. Wang, L. Li, Y. Wu, X. Song, L. Xie, Plasma exosomal tRNA-derived fragments as diagnostic biomarkers in non-small cell lung cancer. Front. Oncol. 12, 1037523 (2022)
J. Xi, Z. Zeng, X. Li, X. Zhang, J. Xu, Expression and diagnostic value of tRNA-Derived fragments secreted by Extracellular vesicles in Hypopharyngeal Carcinoma. Onco Targets Ther. 14, 4189–4199 (2021)
P. Zhang, W. Wu, Q. Chen, M. Chen, Non-coding RNAs and their Integrated Networks. J. Integr. Bioinform 16(2019)
H. Yang, H. Zhang, Z. Chen, Y. Wang, B. Gao, Effects of tRNA-derived fragments and microRNAs regulatory network on pancreatic acinar intracellular trypsinogen activation. Bioengineered. 13, 3207–3220 (2022)
Acknowledgements
The authors are thankful for the Shanghai Science and Technology Committee (No. 20S11901300) and Yunnan Provincial Science and Technology Department - Kunming Medical University Joint Fund Key Project (202201AY070001-137). The authors are thankful for Shanghai University for providing open access support. The authors thank Dr. Yang Shao (Cancer Institute, Fudan University Shanghai Cancer Center) for critical reading of the manuscript.
Funding
This study was funded by the Shanghai Science and Technology Committee (No. 20S11901300) and Yunnan Provincial Science and Technology Department - Kunming Medical University Joint Fund Key Project (202201AY070001-137). Open access funding was provided by Shanghai University.
Author information
Authors and Affiliations
Contributions
Zhongliang Ma, Weiwei Wang, Fan Wu presented conceptualization. Qianqian Yang, Wei Pan, Wei Meng searched literatures. Fan Wu wrote the main manuscript text. Fan Wu, QianqianYang, Wei Pan, Wei Meng prepared figures and tables. Zhongliang Ma, Weiwei Wang supervised the review. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
Not applicable.
Ethics declarations
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, F., Yang, Q., Pan, W. et al. tRNA-derived fragments: mechanism of gene regulation and clinical application in lung cancer. Cell Oncol. 47, 37–54 (2024). https://doi.org/10.1007/s13402-023-00864-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-023-00864-z