Abstract
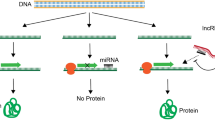
Lung cancer is the utmost familiar category of cancer with greatest fatality rate worldwide and several regulatory mechanisms exercise cellular control on critical oncogenic trails implicated in lung associated carcinogenesis. The non-coding RNAs (ncRNAs) are shown to play a variety of regulatory roles, including stimulating cell proliferation, inhibiting programmed cell death, enhancing cancer cell metastatic ability and acquiring resistance to drugs. Furthermore, ncRNAs exhibit tissue-specific expression as well as great stability in bodily fluids. As a consequence, they are strong contenders for cancer based theragnostics. microRNA (miRNA) alters gene expression primarily by either degrading or interfering with the translation of targeted mRNA and long non-coding RNAs (lncRNAs) can influence gene expression by targeting transcriptional activators or repressors, RNA polymers and even DNA-duplex. lncRNAs are typically found to be dysregulated in lung cancer and hence targeting ncRNAs could be a viable strategy for developing potential therapies as well as for overcoming chemoresistance in lung cancer. The purpose of this review is to elucidate the role of ncRNAs, revisiting the recent studies in lung cancer.

Similar content being viewed by others
Abbreviations
- ncRNA:
-
Non-coding RNA
- miRNA:
-
microRNA
- lncRNAs:
-
Long non-coding RNA
- NSCLC:
-
Non-small cell lung cancer
- mRNA:
-
Messenger RNA
- tRNA:
-
Transfer RNA
- rRNA:
-
Ribosomal RNA
- siRNA:
-
Small interfering RNA
- piRNA:
-
Piwi-interacting RNA
- snoRNA:
-
Small nucleolar RNA
- rRNA:
-
Ribosomal RNA
- SnRNA:
-
Small nuclear RNA
- Endo-SiRNA:
-
Endogenous small-interfering RNA
- SncRNA:
-
Short non-coding RNAs
- SCLC:
-
Small cell lung cancer
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- LCC:
-
Large cell carcinoma
- MALAT1:
-
Metastasis associated in lung adenocarcinoma transcript
- circRNA:
-
Circular RNA
- APAF1:
-
Apoptosis peptidase activating factor 1
- RASA1:
-
RAS P21 protein activator 1
- PDCD4:
-
Programmed cell death 4
- SPRED1/2:
-
Sprouty related EVH1 domain containing 1/2
- SPRY1/3/4:
-
Sprouty 1/3/4
- RBL2:
-
RB Transcriptional corepressor like 2
- M6a:
-
N6 -Methyladenine
- METTL3:
-
Methyltransferase-like 3
- Pri-miRNA:
-
Primary microRNA
- DGCR8:
-
Digeorge syndrome critical region gene 8 microprocessor complex component
- DROSHA:
-
Drosha ribonuclease type II
- BRD4:
-
Bromodomain-containing protein 4
- IRF2:
-
Interferon regulatory factor 2
- SUZ12:
-
SUZ12 polycomb repressive complex 2 subunit
- EZH2:
-
Zeste 2 polycomb repressive complex 2 subunits
- ZFX:
-
Zinc finger X-chromosomal protein
- PTPRU:
-
Protein tyrosine phosphatase receptor type U
- EPAS1:
-
Endothelial PAS domain protein 1
- YBX1:
-
Y-box binding protein 1
- SMAD3:
-
SMAD family member 3
- CTLA4:
-
Cytotoxic T-lymphocyte-associate protein 4
- PD1:
-
Programmed death protein 1
- PD-L1:
-
Programmed death ligand 1
- VHL:
-
Von Hippel-Lindau
- SDH:
-
Succinate dehydrogenase
- FOXO3A:
-
Forkhead box O3
- CXCR4:
-
C-X-C motif chemokine receptor 4
- CAF:
-
Cancer associated fibroblasts
- TME:
-
Tumour microenvironment
- EMT:
-
Epithelial mesenchymal transition
- PDCD4:
-
Programmed cell dealth 4
- APAF1:
-
Apoptosis peptidase activating factor 1
- RASA1:
-
RAS p21 protein activator 1
- SPRED1/2:
-
Sprouty related EVH1 domain containing 1/2
- SPRY1/3/4:
-
Sprouty 1/3/4
- RBL2:
-
RB transcriptional corepressor like 2
- EZH2:
-
Enhancer of zeste 2 polycomb repressive complex 2 subunits
- ZFX:
-
Zinc finger X-chromosomal protein
References
Athie A, Marchese FP, González J, Lozano T, Raimondi I, Juvvuna PK et al (2020) Analysis of copy number alterations reveals the lncRNA ALAL-1 as a regulator of lung cancer immune evasion. J Cell Biol 219:201908078
Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D et al (2014) Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 55:604–614
Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG (2013) catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics 29(22):2928–2930
Anfossi S, Babayan A, Pantel K, Calin GA (2018) Clinical utility of circulating non-coding RNAs—an update. Nat Rev Clin Oncol 15:541–563
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, KuramochiMiyagawa S, Nakano T et al (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207
Bai H, Cao Z, Deng C, Zhou L, Wang C (2012) miR-181a sensitizes resistant leukaemia HL-60/Ara-C cells to Ara-C by inducing apoptosis. J Cancer Res Clin Oncol 138(4):595–602
Bainbridge MN, Warren RL, Hirst M, Romanuik T, Zeng T, Go A, Delaney A, Griffith M, Hickenbotham M, Magrini V et al (2006) Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genom 7:246
Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D (2019) LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res 47(1):D1034–D1037
Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS (2017) Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig New Drugs 35:180–188
Berghmans T, Ameye L, Willems L, Paesmans M, Mascaux C, Lafitte JJ, Meert AP, Scherpereel A, Cortot AB, Cstoth I et al (2013) Identification of microRNA-based signatures for response and survival for non-small cell lung cancer treated with cisplatin-vinorelbine A ELCWP prospective study. Lung Cancer 82:340–345
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366
Billy E, Brondani V, Zhang H, Muller U, Filipowicz W (2001) Specific interference with gene expression induced by long, double stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA 98:14428–14433
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K et al (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99:15524–15529
Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K et al (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295:2492–2502
Chen Q-N, Wei C-C, Wang Z-X, Sun M (2016) Long non-coding RNAs in anticancer drug resistance. Oncotarget 8:1925–1936
Chen Y, Jacamo R, Konopleva M, Garzon R, Croce C, Andreeff M (2013) CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J Clin Investig 123(6):2395–2407
Berghmans T, Ameye L, Lafitte J-J, Colinet B, Cortot A, CsToth I, Holbrechts S, Lecomte J, Mascaux C, Meert A-P et al (2015) Prospective validation obtained in a similar group of patients and with similar high throughput biological tests failed to confirm signatures for prediction of response to chemotherapy and survival in advanced NSCLC: a prospective study from the European Lung Cancer Working Party. Front Oncol 4:386
Chen F-F, Yan J-J, Lai W-W, Jin Y-T, Su I-J (1998) Epstein-barr virus-associated nonsmall cell lung carcinoma. Cancer 82:2334–2342
Dai E, Yang F, Wang J, Zhou X, Song Q, An W, Wang L, Jiang W (2017) ncDR: a comprehensive resource of non-coding RNAs involved in drug resistance. Bioinformatics 33(24):4010–4011
De Bacco F, Luraghi P, Medico E, Reato G, Girolami F, Perera T, Gabriele P, Comoglio PM, Boccaccio C (2011) Induction of MET by ionizing radiation and its role in radio resistance and invasive growth of cancer. J Natl Cancer Inst 103(8):645–661
Delaney G, Jacob S, Featherstone C, Barton M (2005) The role of radiotherapy in cancer treatment: estimating optimal utilization from a review of evidence based clinical guidelines. Cancer-Am Cancer Soc 104(6):1129–1137
Deveson IW, Hardwick SA, Mercer TR, Mattick JS (2017) The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet 33:464–478
Ekert JE, Johnson K, Strake B, Pardinas J, Jarantow S, Perkinson R, Colter DC (2014) Three-dimensional lung tumor microenvironment modulates therapeutic compound responsiveness in vitro—implication for drug development. PLoS ONE 9:e92248
Fan L, Huang C, Li J, Gao T, Lin Z, Yao T (2018) Long non-coding RNA urothelial cancer associated 1 regulates radio resistance via the hexokinase 2/glycolytic pathway in cervical cancer. Int J Mol Med 42:2247–2259
Fang Z, Xu J, Zhang B, Wang W, Liu J, Liang C, Hua J, Meng Q, Yu X, Shi S (2020) The promising role of noncoding RNAs in cancer-associated fibroblasts: an overview of current status and future perspectives. J Hematol Oncol 13(1):154
Feng DD, Zhang H, Zhang P, Zheng YS, Zhang XJ, Han BW, Luo XQ, Xu L, Zhou H, Qu LH, Chen YQ (2011) Down-regulated miR-331-5p and miR-27a are associated with chemotherapy resistance and relapse in leukaemia. J Cell Mol Med 15(10):2164–2175
Fields BD, Nguyen SC, Nir G, Kennedy S (2019) A multiplexed DNA FISH strategy for assessing genome architecture in Caenorhabditis elegans. Elife 8:e42823
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Freeman LA (2013) Cloning full-length transcripts and transcript variants using 5′ and 3′ RACE. Methods Mol Biol 1027:3–17
Gao J, Liu L, Li G, Cai M, Tan C, Han X et al (2019) LncRNA GAS5 confers the radio sensitivity of cervical cancer cells via regulating miR-106b/IER3 axis. Int J Biol Macromol 126:994–1001
Gelali E, Girelli G, Matsumoto M, Wernersson E, Custodio J, Mota A et al (2019) iFISH is a publically available resource enabling versatile DNA FISH to study genome architecture. Nat Commun 10(1):1636
Gómez-Román JJ, Martínez MN, Fernández SL, Val-Bernal JF (2009) Epstein-Barr virus-associated adenocarcinomas and squamous-cell lung carcinomas. Mod Pathol 22:530–537
Giorgetti L, Heard E (2016) Closing the loop: 3C versus DNA FISH. Genome Biol 17(1):215
Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442:199–202
Godinho M, Meijer D, Setyono-Han B, Dorssers LCJ, van Agthoven T (2011) Characterization of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. J Cell Physiol 226:1741–1749
Grivna ST, Beyret E, Wang Z, Lin H (2006) A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20:1709–1714
Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP (2016) Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 166(4):1055–1056
Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–952
Han AJ, Xiong M, Zong YS (2000) Association of Epstein-Barr virus with lymphoepithelioma-like carcinoma of the lung in Southern China. Am J Clin Pathol 114:220–226
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Havelange V, Ranganathan P, Geyer S, Nicolet D, Huang X, Yu X, Volinia S, Kornblau SM, Andreeff M, Croce CM, Marcucci G, Bloomfield CD, Garzon R (2014) Implications of the miR-10 family in chemotherapy response of NPM1- mutated AML. Blood 123(15):2412–2415
He Y, Jing Y, Wei F, Tang Y, Yang L, Luo J et al (2018) Long non-coding RNA PVT1 predicts poor prognosis and induces radio resistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis 9:235
Hickey CJ, Schwind S, Radomska HS, Dorrance AM, Santhanam R, Mishra A, Wu YZ, Alachkar H, Maharry K, Nicolet D, Mrozek K, Walker A, Eiring AM, Whitman SP, Becker H, Perrotti D, Wu LC, Zhao X, Fehniger TA, Vij R, Byrd JC, Blum W, Lee LJ, Caligiuri MA, Bloomfield CD, Garzon R, Marcucci G (2013) Lenalidomide-mediated enhanced translation of C/EBPalpha-p30 protein up-regulates expression of the antileukemic microRNA-181a in acute myeloid leukemia. Blood 121(1):159–169
Ho JC-M, Leung C-C (2018) Management of co-existent tuberculosis and lung cancer. Lung Cancer 122:83–87
Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC (1958) A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem 231:241–257
Hou W, Tian Q, Zheng J, Bonkovsky HL (2010) MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology 51:1494–1504
Hsu MT, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–340
Huttenhofer A, Schattner P, Polacek N (2005) Non-coding RNAs: hope or hype? Trends Genet 21:289–297
Huang H, Chen J, Ding C-M, Jin X, Jia Z-M, Peng J (2018) LncRNA NR2F1-AS1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC1 via miR-363. J Cell Mol Med 22:3238–3245
Kuzembayeva M, Hayes M, Sugden B (2014) Multiple functions are mediated by the miRNAs of Epstein-Barr virus. Curr Opin Virol 7:61–65
Luo ML (2016) Methods to study long noncoding RNA biology in cancer. Adv Exp Med Biol 927:69–107
Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834–838
Movia D, Bazou D, Volkov Y, Prina-Mello A (2018) Multilayered cultures of NSCLC cells grown at the air-liquid interface allow the efficacy testing of inhaled anti-cancer drugs. Sci Rep 8:12920
Lu H, He Y, Lin L, Qi Z, Ma L, Li L et al (2015) Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol 37:1683–1691
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z et al (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855–862
Jena BC, Das CK, Bharadwaj D, Manda M (2020) Cancer associated fibroblast mediated chemoresistance: a paradigm shift in understanding the mechanism of tumor progression. Biochim Biophys Acta (BBA) Rev Cancer 1874(2):188416
Jin C, Yan B, Lu Q, Lin Y, Ma L (2016) The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol 37:4025–4033
Jing L, Yuan W, Ruofan D, Jinjin Y, Haifeng Q (2015) HOTAIR enhanced aggressive biological behaviors and induced radio-resistance via inhibiting p21 in cervical cancer. Tumour Biol 36:3611–3619
Johnson JM, Edwards S, Shoemaker D, Schadt EE (2005) Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet 21:93–102
Kamel LM, Atef DM, Mackawy AMH, Shalaby SM, Abdelraheim N (2019) Circulating long non-coding RNA GAS5 and SOX2OT as potential biomarkers for diagnosis and prognosis of non-small cell lung cancer. Biotechnol Appl Biochem 66:634–642
Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR (2002) Large-scale transcriptional activity in chromosomes 21 and 22. Science 296:916–919
Kapranov P, St Laurent G (2012) Dark matter RNA: existence, function, and controversy. Front Genet 3:60
Kim JH, Nam B, Choi YJ, Kim SY, Lee J-E, Sung KJ, Kim WS, Choi C-M, Chang E-J, Koh JS et al (2018) Enhanced glycolysis supports cell survival in EGFR-mutant lung adenocarcinoma by inhibiting autophagy-mediated EGFR degradation. Cancer Res 78:4482–4496
Kim JS, Kim EJ, Lee S, Tan X, Liu X, Park S, Kang K, Yoon J-S, Ko YH, Kurie JM et al (2019) MiR-34a and miR-34b/c has distinct effects on the suppression of lung adenocarcinomas. Exp Mol Med 51:9
Kim M, Mun H, Sung CO, Cho EJ, Jeon H-J, Chun S-M, Jung DJ, Shin TH, Jeong GS, Kim DK et al (2019) Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun 10:3991
Koshiol J, Gulley ML, Zhao Y, Rubagotti M, Marincola FM, Rotunno M, Tang W, Bergen AW, Bertazzi PA, Roy D et al (2011) Epstein-Barr virus microRNAs and lung cancer. Br J Cancer 105:320–326
Kowalczyk MS, Higgs DR, Gingeras TR (2012) Molecular biology: RNA discrimination. Nature 482:310–311
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16:203–222
Lagarde J, Uszczynska-Ratajczak B, Santoyo-Lopez J, Gonzalez JM, Tapanari E, Mudge JM et al (2016) Extension of humanlncRNA transcripts by RACE coupledwith long-read high-throughput sequencing (RACE-Seq). Nat Commun 7:12339
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294:853–858
Lu Y, Li T, Wei G, Liu L, Chen Q, Xu L et al (2016) The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radio resistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol 37:11733–11741
Ma L, Cao J, Liu L, Du Q, Li Z, Zou D et al (2019) LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res 47(D1):D128–D134
Le P, Romano G, Nana-Sinkam P, Acunzo M (2021) Non-coding RNAs in cancer diagnosis and therapy: focus on lung cancer. Cancers 13(6):1372
Lee SY, Jeong EK, Ju MK, Jeon HM, Kim MY, Kim CH, Park HG, Han SI, Kang HS (2017) Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol Cancer 16(1):10
Lee RC, Feinbaum RL, The AV (1993) C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y et al (2018) A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-17-3454
Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563–574
Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso PT, Guigo R, Johnson R (2017) LncATLAS database for subcellular localization of long noncoding RNAs. RNA 23(7):1080–1087
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S et al (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419
Muhammad II, Kong SL, Akmar AS, Munusamy U (2019) RNA-seq and ChIP-seq as complementary approaches for comprehension of plant transcriptional regulatory mechanism. Int J Mol Sci 21(1):167
Li H, Hui L, Xu W (2012) miR-181a sensitizes a multidrug-resistant leukemia cell line K562/A02 to daunorubicin by targeting BCL-2. Acta Biochim Biophys Sin (Shanghai) 44(3):269–277
Li N, Meng D-D, Gao L, Xu Y, Liu P-J, Tian Y-W et al (2018) Overexpression of HOTAIR leads to radioresistance of human cervical cancer via promoting HIF-1α expression. Radiat Oncol 13:210
Ratan ZA, Zaman SB, Mehta V, Haidere MF, Runa NJ, Akter N (2017) Application of fluorescence in situ hybridization (FISH) technique for the detection of genetic aberration in medical science. Cureus 9(6):e1325
Nakato R, Sakata T (2020) Methods for ChIP-seq analysis: a practical workflow and advanced applications. Methods S1046–2023(20):30059–30061
Pachnis V, Belayew A, Tilghman SM (1984) Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci USA 81:5523–5527
Li S, Mason CE (2014) The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet 15:127–150
Li Y, Ye Y, Feng B, Qi Y (2017) Long noncoding RNA lncARSR promotes doxorubicin resistance in hepatocellular carcinoma via modulating PTENPI3K/Akt pathway. J Cell Biochem 118:4498–4507
Li N, Feng XB, Tan Q, Luo P, Jing W, Zhu M, Liang C, Tu J, Ning Y (2017) Identification of circulating long noncoding RNA Linc00152 as a novel biomarker for diagnosis and monitoring of non-small-cell lung cancer. Dis Markers 2017:7439698
Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB et al (2015) lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res 43(Database issue):D168–D173
Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S, Harrison PM, Nelson FK, Miller P, Gerstein M et al (2003) The transcriptional activity of human chromosome 22. Genes Dev 17:529–540
Rodriguez-Mateos P, Azevedo NF, Almeida C, Pamme N (2020) FISH and chips: a review of microfluidic platforms for FISH analysis. Med Microbiol Immunol 209(3):373–391
Li N, Wang Y, Liu X, Luo P, Jing W, Zhu M, Tu J (2017) Identification of circulating long noncoding RNA HOTAIR as a novel biomarker for diagnosis and monitoring of non-small cell lung cancer. Technol Cancer Res Treat 16:1060–1066
Liang W, Lv T, Shi X, Liu H, Zhu Q, Zeng J, Yang W, Yin J, Song Y (2016) Circulating long noncoding RNA GAS5 is a novel biomarker for the diagnosis of nonsmall cell lung cancer. Medicine 95:e4608
Lim B, Lin Y, Navin N (2020) Advancing cancer research and medicine with single-cell genomics. Cancer Cell 37(4):456–470. https://doi.org/10.1016/j.ccell.2020.03.008
Salgia R (2017) MET in lung cancer: biomarker selection based on scientific rationale. Mol Cancer Ther 16:555–565
Santos RM, Moreno C, Zhang WC (2020) Non-coding RNAs in lung tumor initiation and progression. Int J Mol Sciences 21(8):2774
Sarnow P, Sagan SM (2016) Unraveling the mysterious interactions between hepatitis C virus RNA and liver-specific microRNA-122. Annu Rev Virol 3:309–332
Schwind S, Maharry K, Radmacher MD, Mrozek K, Holland KB, Margeson D, Whitman SP, Hickey C, Becker H, Metzeler KH, Paschka P, Baldus CD, Liu S, Garzon R, Powell BL, Kolitz JE, Carroll AJ, Caligiuri MA, Larson RA, Marcucci G, Bloomfield CD (2010) Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol 28(36):5257–5264
Seca H, Lima RT, Almeida GM, Sobrinho-Simoes M, Bergantim R, Guimaraes JE, Vasconcelos MH (2014) Effect of miR-128 in DNA damage of HL-60 acute myeloid leukemia cells. Curr Pharm Biotechnol 15(5):492–502
Liu F, Zheng K, Chen HC, Liu ZF (2018) Capping-RACE: a simple, accurate, and sensitive 5′ RACE method for use in prokaryotes. Nucleic Acids Res 46(21):e129
Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H et al (2009) MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA 106:12085–12090
Shen H, Shen J, Wang L, Shi Z, Wang M, Jiang B-H, Shu Y (2015) Low miR-145 expression level is associated with poor pathological differentiation and poor prognosis in non-small cell lung cancer. Biomed Pharmacother 69:301–305
Sia D, Villanueva A, Friedman SL, Llovet JM (2017) Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology 152:745–761
Slack FJ, Chinnaiyan AM (2019) The role of non-coding RNAs in oncology. Cell 179:1033–1055
Stark R, Grzelak M, Hadfield J (2019) RNA sequencing: the teenage years. Nat Rev Genet 20:631–656
Wang S, Xiong H, Yan S, Wu N, Lu Z (2016) Identification and characterization of Epstein-Barr virus genomes in lung carcinoma biopsy samples by next-generation sequencing technology. Sci Rep 6:26156
Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H (2006) Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20:1732–1743
Stenvold H, Donnem T, Andersen S, Al-Saad S, Busund L-T, Bremnes RM (2014) Stage and tissue-specific prognostic impact of miR-182 in NSCLC. BMC Cancer 14:138
Teixeira FK, Okuniewska M, Malone CD, Coux R-X, Rio DC, Lehmann R (2017) piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552:268–272
Tomczak K, Czerwinska P, Wiznerowicz M (2015) The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 19(1A):A68-77
Topol EJ (2019) High-performance medicine: the convergence of human and artificial intelligence. Nat Med 25:44–56
Van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, Huynh Y, Chrzanowska A, Fulham MJ, Bailey DL et al (2017) Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol 18:1386–1396
Vizzini A (2020) 5′ and 3′ RACE method to obtain full-length 5′ and 3′ ends of Ciona robusta macrophage migration inhibitory factors Mif1 and Mif2 cDNA. Methods Mol Biol 2080:223–235
Wang H, Li Q, Tang S, Li M, Feng A, Qin L, Liu Z, Wang X (2017) The role of long noncoding RNA HOTAIR in the acquired multidrug resistance to imatinib in chronic myeloid leukemia cells. Hematology 22(4):208–216
Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL (2011) Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol 30:99–104
Migliore C, Morando E, Ghiso E, Anastasi S, Leoni VP, Apicella M, Cora D, Sapino A, Pietrantonio F, De Braud F et al (2018) miR-205 mediates adaptive resistance to MET inhibition via ERRFI1 targeting and raised EGFR signaling. EMBO Mol Med 10:e8746
Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, Heo I, Böttinger L, Klay D, Weeber F, Huelsz-Prince G, Iakobachvili N, Amatngalim GD et al (2019) Long-term expanding human airway organoids or disease modeling. EMBO J 38:e100300
Xiao J, Lv Y, Jin F, Liu Y, Ma Y, Xiong Y et al (2017) Lncrna HANR promotes tumorigenesis and increase of chemoresistance in hepatocellular carcinoma. Cell Physiol Biochem 43:1926–1938
Wei L, Wang X, Lv L, Liu J, Xing H, Song Y et al (2019) The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer 18:147
Runowicz CD, Leach CR, Henry NL, Henry KS, Mackey HT, Cowens-Alvarado RL et al (2016) American cancer society/American society of clinical oncology breast cancer survivorship care guideline. CA Cancer J Clin 66:43–73
Funding
This study was not funded by any authority.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Roy, S., Ganguly, N. & Banerjee, S. Exploring clinical implications and role of non-coding RNAs in lung carcinogenesis. Mol Biol Rep 49, 6871–6883 (2022). https://doi.org/10.1007/s11033-022-07159-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07159-w