Abstract
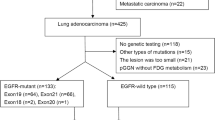
The purpose of this study is to develop the predictive models for epidermal growth factor receptor (EGFR) mutation status and subtypes [exon 21-point mutation (L858R) and exon 19 deletion mutation (19Del)] and evaluate their clinical usefulness. Total 172 patients with lung adenocarcinoma were retrospectively analyzed. The analysis of variance and the least absolute shrinkage were used for feature selection from plain computed tomography images. Then, radiomic score (rad-score) was calculated for the training and test cohorts. Two machine learning (ML) models with 5-fold were applied to construct the predictive models with rad-score, clinical features, and the combination of rad-score and clinical features. The nomogram was developed using rad-score and clinical features. The prediction performance was evaluated by the area under the receiver operating characteristic curve (AUC). Finally, decision curve analysis (DCA) was performed using the best ML and nomogram models. In the test cohorts, the AUC of the best ML and the nomogram model were 0.73 (95% confidence interval, 0.59–0.87) and 0.79 (0.65–0.92) in the EGFR mutation groups, 0.83 (0.67–0.99) and 0.85 (0.72–0.97) in the L858R mutation groups, as well as 0.77 (0.58–0.97) and 0.77 (0.60–0.95) in the 19Del groups. The DCA showed that the nomogram models have comparable results with ML models. We constructed two predictive models for EGFR mutation status and subtypes. The nomogram models had comparable results to the ML models. Because the superiority of the performance of ML and nomogram models varied depending on the prediction groups, appropriate model selection is necessary.



Similar content being viewed by others
References
Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG et al (2011) Non-small-cell lung cancer. Lancet 378:1727–1740. https://doi.org/10.1016/S0140-6736(10)62101-0
Rizzo S, Petrella F, Buscarino V, De Maria F, Raimondi S, Barberis M et al (2016) CT radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer. Eur Radiol 26:32–42. https://doi.org/10.1007/s00330-015-3814-0
Mitsudomi T, Yatabe Y (2007) Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci 98:1817–1824. https://doi.org/10.1111/j.1349-7006.2007.00607.x
Jänne PA, Engelman JA, Johnson BE (2005) Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol 23:3227–3234. https://doi.org/10.1200/JCO.2005.09.985
Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H et al (2010) Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N Engl J Med 362:2380–2388. https://doi.org/10.1056/nejmoa0909530
Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J et al (2010) Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 11:121–128. https://doi.org/10.1016/S1470-2045(09)70364-X
Kriegs M, Gurtner K, Can Y, Brammer I, Rieckmann T, Oertel R et al (2015) Radiosensitization of NSCLC cells by EGFR inhibition is the result of an enhanced p53-dependent G1 arrest. Radiother Oncol 115:120–127. https://doi.org/10.1016/j.radonc.2015.02.018
Li S, Luo T, Ding C, Huang Q, Guan Z, Zhang H (2020) Detailed identification of epidermal growth factor receptor mutations in lung adenocarcinoma: combining radiomics with machine learning. Med Phys 47:3458–3466. https://doi.org/10.1002/mp.14238
Soria JC, Mok TS, Cappuzzo F, Jänne PA (2012) EGFR-mutated oncogene-addicted non-small cell lung cancer: current trends and future prospects. Cancer Treat Rev 38:416–430. https://doi.org/10.1016/j.ctrv.2011.10.003
Lee CK, Davies L, Wu YL, Mitsudomi T, Inoue A, Rosell R et al (2017) Gefitinib or erlotinib vs chemotherapy for EGFR mutation-positive lung cancer: individual patient data meta-analysis of overall survival. J Natl Cancer Inst 109:1–9. https://doi.org/10.1093/jnci/djw279
Sutiman N, Tan SW, Tan EH, Lim WT, Kanesvaran R, Ng QS et al (2017) EGFR mutation subtypes influence survival outcomes following first-line gefitinib therapy in advanced asian NSCLC patients. J Thorac Oncol 12:529–538. https://doi.org/10.1016/j.jtho.2016.11.2225
Liu Q, Sun D, Li N, Kim J, Feng D, Huang G et al (2020) Predicting EGFR mutation subtypes in lung adenocarcinoma using 18F-FDG PET/CT radiomic features. Transl Lung Cancer Res 9:549–562. https://doi.org/10.21037/tlcr.2020.04.17
Zhang L, Zhang Y, Chang L, Yang Y, Fang W, Guan Y et al (2019) Intratumor heterogeneity comparison among different subtypes of non-small-cell lung cancer through multi-region tissue and matched ctDNA sequencing. Mol Cancer 18:1–6. https://doi.org/10.1186/s12943-019-0939-9
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577. https://doi.org/10.1148/radiol.2015151169
Park H, Sholl LM, Hatabu H, Awad MM, Nishino M (2019) Imaging of precision therapy for lung cancer: current state of the art. Radiology 293:15–29. https://doi.org/10.1148/radiol.2019190173
Li S, Ding C, Zhang H, Song J, Wu L (2019) Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med Phys 46:4545–4552. https://doi.org/10.1002/mp.13747
Hong D, Xu K, Zhang L, Wan X, Guo Y (2020) Radiomics signature as a predictive factor for EGFR mutations in advanced lung adenocarcinoma. Front Oncol 10:1–8. https://doi.org/10.3389/fonc.2020.00028
Mei D, Luo Y, Wang Y, Gong J (2018) CT texture analysis of lung adenocarcinoma: can Radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging 18:1–9. https://doi.org/10.1186/s40644-018-0184-2
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, Van Stiphout RGPM, Granton P et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446. https://doi.org/10.1016/j.ejca.2011.11.036
Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Cavalho S et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:1–9. https://doi.org/10.1038/ncomms5006
Zhao W, Wu Y, Xu Y, Sun Y, Gao P, Tan M et al (2020) The potential of radiomics nomogram in non-invasively prediction of epidermal growth factor receptor mutation status and subtypes in lung adenocarcinoma. Front Oncol 9:1485. https://doi.org/10.3389/fonc.2019.01485
Velazquez ER, Parmar C, Jermoumi M, Mak RH, Van Baardwijk A, Fennessy FM et al (2013) Volumetric CT-based segmentation of NSCLC using 3D-slicer. Sci Rep 3:1–7. https://doi.org/10.1038/srep03529
Van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Tixier F, Le Rest CC, Hatt M, Albarghach N, Pradier O, Metges JP et al (2011) Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med 52:369–378. https://doi.org/10.2967/jnumed.110.082404
Shiinoki T, Fujimoto K, Kawazoe Y, Yuasa Y, Kajima M, Manabe Y et al (2022) Predicting programmed death-ligand 1 expression level in non-small cell lung cancer using a combination of peritumoral and intratumoral radiomic features on computed tomography. Biomed Phys Eng Express 8:25008. https://doi.org/10.1088/2057-1976/ac4d43
Xiong Z, Jiang Y, Tian D, Zhang J, Guo Y, Li G et al (2022) Radiomics for identifying lung adenocarcinomas with predominant lepidic growth manifesting as large pure ground-glass nodules on CT images. PLoS ONE 17:1–15. https://doi.org/10.1371/journal.pone.0269356
Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z et al (2016) Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology 281:947–957. https://doi.org/10.1148/radiol.2016152234
Kuang Y, Li R, Jia P, Ye W, Zhou R, Zhu R et al (2021) MRI-Based Radiomics: Nomograms predicting the short-term response after transcatheter arterial chemoembolization (TACE) in hepatocellular carcinoma patients with diameter less than 5 cm. Abdom Radiol 46:3772–3789. https://doi.org/10.1007/s00261-021-02992-2
Li X, Yin G, Zhang Y, Dai D, Liu J, Chen P et al (2019) Predictive power of a Radiomic signature based on 18F-FDG PET/CT images for EGFR Mutational Status in NSCLC. Front Oncol 9:1–11. https://doi.org/10.3389/fonc.2019.01062
Cho A, Hur J, Moon YW, Hong SR, Suh YJ, Kim YJ et al (2016) Correlation between EGFR gene mutation, cytologic tumor markers, 18F-FDG uptake in non-small cell lung cancer. BMC Cancer 16:1–8. https://doi.org/10.1186/s12885-016-2251-z
Zhang B, Liu Q, Zhang X, Liu S, Chen W, You J et al (2020) Clinical utility of a Nomogram for Predicting 30-Days poor outcome in hospitalized patients with COVID-19: Multicenter External validation and decision curve analysis. Front Med 7:1–12. https://doi.org/10.3389/fmed.2020.590460
Acknowledgements
We thank Medical Physics Research Unit in Yamaguchi University (https://ds0n.cc.yamaguchi-u.ac.jp/~medphys/) for providing support to accomplish this study.
Funding
This study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant number 20K16789 (KF), 22K07667 (TS), and the Takeda Science Foundation.
Author information
Authors and Affiliations
Contributions
YK and TS carried out the experiment. YK wrote the manuscript with support from TS, KF, YY. TH and KM supplied available data in terms of this study and discussed. HT supervised this study. All authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the Institutional Review Board of Yamaguchi University, Japan. The ethics certificate number was #2020−148.
Consent to participants.
This study was retrospective study. We applied opt-out method to obtain consent on this study. The opt-out method was approved by the Institutional Review Board of Yamaguchi University.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kawazoe, Y., Shiinoki, T., Fujimoto, K. et al. Comparison of the radiomics-based predictive models using machine learning and nomogram for epidermal growth factor receptor mutation status and subtypes in lung adenocarcinoma. Phys Eng Sci Med 46, 395–403 (2023). https://doi.org/10.1007/s13246-023-01232-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-023-01232-9