Abstract
The epidermal growth factor receptor (EGFR) Thr790 Met (T790M) mutation is responsible for approximately half of the acquired resistance to EGFR-tyrosine kinase inhibitor (TKI) in non-small-cell lung cancer (NSCLC) patients. Identifying patients at diagnosis who are likely to develop this mutation after first- or second-generation EGFR-TKI treatment is crucial for better treatment outcomes. This study aims to develop and validate a radiomics-based machine learning (ML) approach to predict the T790M mutation in NSCLC patients at diagnosis. We collected retrospective data from 210 positive EGFR mutation NSCLC patients, extracting 1316 radiomics features from CT images. Using the LASSO algorithm, we selected 10 radiomics features and 2 clinical features most relevant to the mutations. We built models with 7 ML approaches and assessed their performance through the receiver operating characteristic (ROC) curve. The radiomics model and combined model, which integrated radiomics features and relevant clinical factors, achieved an area under the curve (AUC) of 0.80 (95% confidence interval [CI] 0.79–0.81) and 0.86 (0.87–0.88), respectively, in predicting the T790M mutation. Our study presents a convenient and noninvasive radiomics-based ML model for predicting this mutation at the time of diagnosis, aiding in targeted treatment planning for NSCLC patients with EGFR mutations.
Similar content being viewed by others
Introduction
Lung cancer is one of the most commonly diagnosed cancers and the leading cause of cancer-related death worldwide1. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of primary lung cancer2. Although considerable progress has been achieved in the treatment of NSCLC over the past two decades, the overall cure and survival rates for NSCLC remain low, particularly in patients with advanced/metastatic disease3.
Epidermal growth factor receptor (EGFR) mutations are identified in approximately 20% of patients with NSCLC in the Caucasian population, and up to 40% in the Asian population4,5. First-generation (gefitinib, erlotinib) and second-generation (afatinib, dacomitinib) EGFR-tyrosine kinase inhibitors (TKIs) serve as standard first-line therapy for treatment-naive patients with sensitizing EGFR mutation-positive advanced/metastatic NSCLC patients because these TKIs have provided patients with clinical benefit, such as high response rate and prolonged progression-free survival (PFS) versus platinum-based doublet chemotherapy6,7,8,9,10. However, most patients develop resistance within 10–14 months after initial treatment11.
The most frequently identified mechanism of acquired TKI resistance is an EGFR Thr790Met (T790M) point mutation within exon 20, which confers drug resistance by increasing ATP affinity12. Osimertinib, a third-generation EGFR-TKI which selectively inhibits both EGFR sensitizing mutations and EGFR T790M resistance mutations, had significantly greater efficacy than platinum-based doublet chemotherapy in T790M-positive advanced NSCLC patients who had progressed during first-line EGFR-TKI therapy13. At present, osimertinib is a treatment choice for patients with EGFR mutation-positive advanced NSCLC in the first-line setting, and for patients with T790M positive NSCLC following disease progression after first-line EGFR-TKIs12,14.
Although 3 generations of EGFR-TKIs are currently available for the treatment of EGFR mutation-positive NSCLC, the optimal sequence of administrating these drugs to maximize the duration of the EGFR signaling inhibition remains still uncertain15,16. For patients who are likely to develop the EGFR T790M mutation, a sequencing strategy of the first- or second-generation TKIs followed by osimertinib has shown promising outcomes16,17. For T790M-negative patients, osimertinib may be a first-choice TKI14,18. Therefore, it is of great importance to identify patients at the time of diagnosis who would be likely to acquire T790M after treatment with a first- or second-generation EGFR-TKI, as this will enable appropriate screening for improved treatment outcomes.
Radiomics is a rapidly evolving field related to the computerized extraction and analysis of data from digital medical images, which offers unique potential to significantly improve the efficiency and accuracy of lung cancer screening, as well as enhance clinical decision-making19,20,21. By extracting imaging information from magnetic resonance imaging (MRI), computed tomography (CT), and positron-emission-tomography (PET), radiomics analysis can be performed to characterize histology and genotype of nodules, identify patient candidate for molecular targeted therapy and immunotherapy, predict treatment response and potential side effects of radiation and immunotherapy, and even differentiate lung injury from recurrence22,23,24,25,26. In a previous study, we built a model integrating radiomics and clinical variables for prediction of EGFR mutation and achieved an AUC of 0.8627. Yang and colleague collected thoracic CT scans from patients who had confirmed progression on first- or second-generation TKIs and developed a model based on radiomics features and clinical data to detect acquired T790M mutation28. They reported an AUC of 0.71 for predicting T790M mutation in patients with advanced lung adenocarcinoma who experienced progression after first- or second-generation EGFR-TKI therapy. However, it is even more important to identify acquired T790M mutation in untreated NSCLC patients in order to optimize the sequence of EGFR-TKI administration. Yet, to date, very few studies have investigated whether radiomics may be capable of predicting the likelihood of developing T790M in treatment-naïve NSCLC patients after a first- or second-generation EGFR-TKI therapy. Therefore, the aim of this study is to establish a radiomics-based model for predicting acquired EGFR T790M mutation in patients with advanced or metastatic NSCLC harbouring an EGFR-activating mutation.
Material and methods
Patients
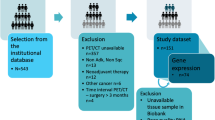
The study population was retrospectively selected from patients with NSCLC at the First Affiliated Hospital of Shandong First Medical University (Jinan, China) between Jan. 2018 and Dec. 2021. This study received approval from the institutional review board of the First Affiliated Hospital of Shandong First Medical University, with a waiver for the requirement of informed consent. Patients who were: (1) histologically diagnosed with primary NSCLC, (2) with known EGFR sensitive mutation, (3) treatment-naïve subjects at the time of diagnosis of NSCLC, (4) classified as unresectable stage III or metastatic (stage IV) according to the Eighth Edition of the Lung Cancer Stage Classification29, (5) receiving chest CT scan prior to biopsies, (6) treated with either gefitinib, erlotinib, icotinib, or afatinib as first-line EGFR-TKI therapy, (7) receiving re-biopsy after TKI failure and were tested for EGFR T790M mutation, met the inclusion criteria and were included. The exclusion criteria were given as follows: (1) lack of clinical and demographic data, such as age, gender, smoking status, stage, and levels of serum tumor marker, (2) difficulty in drawing regions of interest (ROIs), (3) poor quality of CT images. In the end, 274 patients were included and randomly assigned to a training cohort (n = 192) and a validation cohort (n = 82). The detailed process of screening and grouping of NSCLC cases is shown in Supplementary Fig. 1.
Analysis of EGFR Mutation
The tumor specimen was examined at diagnosis for EGFR mutations in exons 18, 19, 20, and 21 by an amplification refractory mutation system (ARMS) real-time technology using Human EGFR Gene Mutations Fluorescence Polymerase Chain Reaction (PCR) Diagnostic Kit (Amoy Diagnostics Co., Ltd, Xiamen, China) or next-generation sequencing (NGS) (Xiansheng Medical Diagnosis Co., Ltd, Nanjing, China). The presence of T790M mutation was detected on relapsed tumor tissue or circulating free DNA (cfDNA) from plasma sample also by ARMS-PCR or NGS, as described elsewhere30,31.
Image acquisition
All CT scans were performed prior to any treatment for lung cancer using two CT scanners (GE Healthcare, Milwaukee, WI, USA; United Imaging, Shanghai, China). The scanning parameters were as follows: the tube voltage, 120 kVp; tube current, 160–300 mA; detector collimation, 64 or 128 × 0.625 mm; field of view, 350 × 350 mm; the pitch, 0.992:1; and matrix of 512 × 512. All images were reconstructed with a section thickness of 2 mm and subsequently stored in DICOM format in the Picture Archiving and Communication Systems (PACS) using mediastinal (width, 360 HU; level, 50 HU) and lung (width, 1500 HU; level, -650 HU) window settings.
Image preprocessing
Due to the use of different CT scans in the present study, image preprocessing prior to segmentation and feature extraction was performed to make the radiomic features more robust and more suitable for further analysis32. A resampling method was used in this process according to the modified protocol reported previously33. In brief, the CT image pixel values were first converted from radiodensity to Hounsfield Units (HU) using the metadata attributes of the scans. Then, the entire dataset, including tumor masks, was resampled to standardize image representations. The spacing between slices and pixel spacing were set to 1 mm and [1.0, 1.0] mm, respectively. Each slice dimension was adjusted accordingly to match the new spacing, and the resampled image was obtained through interpolation.
Tumor segmentation
Tumor regions of interest (ROI) were manually segmented slice by slice by a senior radiologist with over 10-year experience of CT interpretation using ITK-SNAP (Version 3.6, www.itksnap.org)34,35. The ROI segmentation was subsequently verified by another chest radiologist with 15-year experience. Both radiologists had no knowledge of the clinical data and mutational status. When one patient has multiple lesions, the radiologist only delineates the tumor area where the biopsy was performed. To assess the consistency of segmentation between the two radiologists, the intra-group correlation coefficient (ICC) for each feature was calculated36,37. Only features with an ICC greater than 0.85 were deemed to have high stability and were remained for the further analysis.
Radiomic feature extraction
In this study, radiomics features were extracted from each three-dimensional (3D) ROI using Pyradiomics library (http://pyradiomics.readthedocs.io/en/latest/index.html), which was in conformance with the Image Biomarker Standardization Initiative 38. These features can be classified as 3 categories: first-order statistics, shape-based, and textural feature39. The textural feature category consists of gray level co-occurrence matrix (GLCM), gray level run length matrix (GLRLM), gray level size zone matrix (GLSZM), gray level dependence matrix (GLDM), and neighboring gray tone difference matrix (NGTDM). Moreover, two filters (including wavelet and Laplacian of Gaussian were also utilized to obtain transformed images from the original CT images. By decomposing the image with wavelet transform, high (H) or low (L) pass filter in 3D were used and 8 kinds of combinations were obtained: LHL, HHL, HLL, HHH, HLH, LHH, LLH, and LLL. To emphasize areas of gray level change, the LoG filter was applied to the input image and yield a derived image for each sigma value specified40. Five filters with different sigma values (sigma = 1.0 mm, 2.0 mm, 3.0 mm, 4.0 mm, 5.0 mm) were applied in the present study.
Radiomic feature selection
Although numerous radiomic features were extracted and quantified, not all of them showed an association with the status of T790M mutation. Therefore, feature selection is of great importance to improving the generalization ability and optimizing the model. In this study, the z-score method was initially used to standardize all radiomics features in order to reduce the redundancy between these features. Subsequently, the Wilcoxon rank sum test was used to retain features with a P-value < 0.0541. Then, the least absolute shrinkage and selection operator (LASSO) algorithm, which was suitable for high-dimensional, small-sample size data with the problem of collinearity42,43, was employed to select potential features related to T790M mutational status. LASSO can identify the most predictive features while minimizing overfitting and selection bias. Finally, backward stepwise logistic regression analysis was conducted to select the variables for model building. The termination rule for this process was based on the likelihood ratio test with Akaike’s information criterion (AIC) minimum method44.
Models establishment and performance evaluation
After feature selection, 7 machine learning (ML) methods were imported from the scikit-learn library in Python software to construct models45. These algorithms included decision tree (DT), k nearest neighbors (KNN), logistic regression (LR), naïve Bayes (NB), random forest (RF), support vector machines (SVM), and extreme gradient boosting (XGBoost, XGB). The performances of these models were first assessed by analysis of area under the curve (AUC) of receiver operating characteristic (ROC) curve, sensitivity, and specificity in the validation cohort. Fivefold cross-validation was simultaneously applied to evaluate all results. Then, the optimal model was selected for further analysis.
Several clinical features are associated with mutant EGFR status in NSCLC patients46,47. Therefore, some clinical factors were included in the analysis. They consisted of age, gender, smoking status, performance status (at biopsy), stage of disease, serum level of tumor markers, and the initial response to first- or second-generation EGFR-TKI. The Chi-square and Student’s t-tests were first used to screen clinical factors related to EGFR T790M mutation and those with a P-value lower than 0.05 were retained for further analysis.
To enhance the prediction accuracy even further, the optimal classifier and significant clinical features correlated to EGFR T790M mutation were integrated to establish the combined models. The predictive performances of these models were also evaluated based on the AUC of ROC curve analysis.
Nomogram construction
Nomograms utilize multiple prognostic and determinant variables to establish a statistical prognostic model which can generate an individual probability of a clinical event, thus aiding clinical decision making48. In the present study, a nomogram model was developed based on a multivariable logistic analysis. For each patient, a radiomics score (Rad-score) was calculated by assigning weights to discriminating radiomic features based on their respective coefficients. The radiomics signature and clinical factors were incorporated into a nomogram model designed to predict EGFR T790M mutation in the training cohort. The predictive accuracy of the model was evaluated by a calibration curve. Decision curve analysis (DCA) was conducted to examine the performance of a model to predict EGFR T790M mutation by quantifying the net benefits of 2 models in both the training and validation cohorts33. The workflow of the radiomic analysis is depicted in Fig. 1.
Statistical analysis
Statistical analysis was conducted using PRISM version 6 (GraphPad, La Jolla, CA, USA). Quantitative data were compared using Student’s t-test, and categorical data were compared using the χ2 test to identify any baseline differences. The discrimination performance of models was assessed by the ROC curve and the AUC, sensitivity, and specificity of the model were calculated. All statistical tests were two-tailed, and statistical significance was set a priori at 0.05.
Ethics statement
The study was reviewed and approved by the Institutional Review Committee of the First Affiliated Hospital of Shandong First Medical University (Jinan, China) (approval no. 2019-S-306). Written informed consent for participation is not required for this study in accordance with national legislative and institutional requirements.
Results
Clinical characteristics of patients
The baseline clinical characteristics of the enrolled patients were shown in Table 1. No evident differences were found with respect to the age, smoking status , performance status (at biopsy), stage of disease, pathological type, EGFR mutation status, and serum level of CEA, NSE, CYFRA 21-1, and Pro-GRP between patients with the EGFR T790M mutation and those without (P > 0.05). However, a substantial difference was observed in the initial response to first- or second-generation EGFR-TKI between T790M-positive and T790M-negative patients. In both the training cohort and the validation cohort, the objective remission rate (ORR) in the T790M-positive group was significantly higher than that in the T790M-negative group (P < 0.05). In addition, a higher proportion of male individuals were observed in T790M-positive patients compared to T790M-negative patients in both the training cohort and the validation cohort (P < 0.05).
Feature extraction and selection
In total, 1316 radiomic features were successfully extracted from each patient’s ROI. Clinical features included gender and initial response to first- or second-generation EGFR-TKI. The ICC (= mean ± SD) for radiomics features in each group were calculated. Shape-based features (ICC = 0.92 ± 0.07), first-order statistics (ICC = 0.93 ± 0.03), GLRLM-derived texture features (ICC = 0.96 ± 0.04), GLCM-derived texture features (ICC = 0.97 ± 0.02), met the criteria for high stability (ICC > 0.85), while GLSZM-derived texture features (ICC = 0.56 ± 0.44), GLDM-derived texture features (ICC = 0.36 ± 0.44), NGTDM-derived texture features (ICC = 0.59 ± 0.32), LoG features (ICC = 0.77 ± 0.35), and wavelet (ICC = 0.78 ± 0.36) features did not. Out of the initial 1316 features, 955 (72.60%) were identified as stable and retained. These features comprised 13 shape-based features, 18 first-order features, 16 GLRLM features, 24 GLCM features, 6 GLSZM features, 5 GLDM features, 3 NGTDM features, 544 wavelet transformed features, and 326 LoG transformed features. The histogram of the ICC values of the radiomic features was shown in Supplementary Fig. 2.
Subsequently, the dimensionality reduction was performed and the coefficient for each selected feature was illustrated in Fig. 2. As shown in Fig. 2B,C, when the variable is equal to 10, the error classification value is lower. Thus, the 10 features were selected to build the LASSO logistic regression model. The coefficients for each selected radiomics feature are shown in Fig. 2D. Figure 3 shows the 10 radiomics features were significantly different between the T790M-positive and T790M-negative patients in the training set.
Radiomics feature selection process. (A) The features were screened using the Wilcoxon rank-sum test, and the test level was 0.05. (B, C) The LASSO was used to further filter the most relevant features. (D) Coefficients for each selected radiomics feature. MSE: mean square error, LASSO: least absolute shrinkage and selection operator.
Predictive performance assessment of models
The predictive performance of all 7 models based on radiomics was presented in Supplementary Fig. 3. Among these models, the ML method of RF exhibited superior performance compared to the others in the validation cohorts. The parameters of the RF model were set as follows: the number of trees was 2, and the maximum depth was 4 (Supplementary Fig. 4). In terms of predicting EGFR T790M mutation, the RF and combined models obtained an AUC, sensitivity, specificity, and accuracy of 0.80 (95% confidence interval [CI] 0.79–0.81), 0.86 (0.84–0.89); 0.85 (0.81–0.89), 0.78 (0.72–0.84); 0.70 (0.65–0.74), 0.76 (0.67–0.85); and 0.75 (0.71–0.78), 0.77 (0.73–0.82); respectively using fivefold cross-validation (Fig. 4A,B). Moreover, the combined model, which integrated the radiomics signature derived from RF model and the clinical factors, demonstrated even higher AUC values in the training and validation cohorts: 0.92 and 0.87, respectively Fig. 4C,D.
Analysis of the radiomic nomogram
The radiomic nomogram model developed by integrating the radiomics signatures derived from the RF classifier and clinical predictors achieved higher AUC values in the validation cohort [0.86 (95% CI 0.85–0.88)] (Fig. 5). These findings indicated that the combined model exhibited enhanced predictive capabilities for predicting EGFR T790M mutation. To assess the predictive ability of the model, a calibration curve was plotted. The DCA for the radiomics nomogram and is presented in Fig. 6. Remarkably, the combined model outperformed the other models, as evidenced by its larger area under the decision curve, signifying its superior clinical utility.
Discussion
In our current study, we built a ML-derived radiomics model utilizing non-invasive CT images to predict EGFR T790M mutation in treatment-naïve patients with NSCLC. This model achieved promising performance in the validation cohort (AUC 0.80, 95% CI 0.79–0.81). In particular, the sensitivity and specificity to identify T790M mutation was 0.85 (0.81–0.89), and 0.70 (0.65–0.74), respectively, indicating a low false-positive and false-negative rate, which would be of help in accurately screening patients with EGFR T790M mutation. Moreover, we proposed a predictive model integrating CT images and clinical features, which achieved an AUC of 0.86 (95% CI 0.81–0.89), a sensitivity of 0.78 (0.72–0.84), and a specificity of 0.76 (0.67–0.85). This model further demonstrated the ability to identify acquired EGFR T790M mutation in patients who received first- or second-generation EGFR-TKIs. Our study unveils the potential of a noninvasive approach in identifying patients who are likely to develop the T790M mutation following treatment with first- and second-generation EGFR-TKIs. Importantly, this is the first study, to the best of our knowledge, investigating the ability of radiomics-based models to predict acquired EGFR T790M mutation in Chinese patients with advanced NSCLC bearing an EGFR-activating mutation by utilizing CT imaging from treatment-naïve patients with NSCLC. Our findings support the feasibility of using this model to identify EGFR T790M mutation, offering valuable guidance in selecting appropriate patients for improved treatment outcomes. This study represents a significant contribution to the field of radiomics research, providing novel insights and potential advancements.
Radiomics is a newly emerging and rapidly progressing field that integrates radiology, oncology, and ML techniques49. By utilizing radiomics data, descriptive and predictive models can be developed, providing invaluable diagnostic, prognostic, or predictive information50. Moreover, certain radiomics features are even able to detect genomic alterations within tumor DNA, leading to the emergence of ‘radiogenomics’20. Although some limitations of the radiogenomic approach may exist, radiogenomics is playing an increasingly important role in precision diagnostics and optimal therapy in lung cancer. For example, radiogenomics can be of help to treatment option and prognosis assessment in NSCLC patients51,52. In addition, radiogenomics can aid in evaluating efficacy of therapy and predicting outcomes of patients51,52. Therefore, radiogenomics holds great promise for improving decision making, facilitating more precise personalized care, and ultimately improving patient outcomes.
The findings of this study concur with a previous study that highlighted the correlation between radiomics features from baseline chest CT and the subsequent development of the T790M in Caucasian patients with NSCLC after treatment with an EGFR inhibitor24. Kim et al. also reported that smaller tumor size and selection of metastatic lung lesions as biopsy targets were associated with the detection of the T790M mutation at re-biopsy for mutational analysis53. Furthermore, significant differences in CT imaging were observed between acquired and primary T790M mutations54. Our results, combined with previous research, clearly demonstrate the feasibility of identifying individuals who are likely to acquire the T790M mutation after receiving first- or second-generation EGFR-TKIs using baseline CT scans. Intriguingly, Koo et al.55 retrospectively analyzed CT findings of NSCLC patients at the initial diagnosis and those at re-biopsy and found that peripheral tumor location with vascular convergence, the presence of a pleural tag, and air bronchogram at the time of re-biopsy were associated with acquired T790M mutation. Yoshida and colleagues56 utilized PET scans and found that patients with T790M mutation exhibited lower levels of 18F-2-fluoro-2-deoxyglucose uptake in comparison with those without T790M mutation. These findings further validate the link between radiomics features and development of T790M during treatment with EGFR-TKI. Furthermore, models that integrate both radiomic features and clinical factors demonstrated excellent performance in evaluating the prognosis of metastatic NSCLC patients with EGFR-T790M mutation receiving osimertinib treatment57.
When some clinical characteristics added to the radiomics based model, the ability of the combined model to predict the T790M mutation has also been assessed in the present study. The combined model, composed of clinicopathologic and CT-radiomic signatures, achieved good detection performance with an AUC 0.87 in the validation datasets. These findings suggests that certain clinical features may contribute to the discrimination of the T790M mutation within the combined model. We identified gender and initial response to first- or second-generation EGFR-TKI as the most influential clinical predictor. These results are consistent with previous studies that reported that gender (male), initial EGFR-TKI response (complete or partial response), progression pattern (solitary lesion progression), longer duration of EGFR-TKI, postsurgery recurrence, may represent useful predictive markers for T790M detection58,59. Additionally, Dal Maso et al. demonstrated a correlation between age, type of EGFR mutation at diagnosis, response to first-line treatment, andT790M status60. A study by Hou et al. compared the clinical and CT imaging characteristics between primary and acquired EGFR T790M mutations in treatment-naïve patients with NSCLC. They found that patients with primary T790M mutation exhibited earlier tumor stage, higher differentiation, and proportion of lepidic subtype adenocarcinoma in comparison to those with acquired T790M mutations54.
Liquid biopsy, which involves analyzing circulating tumor-derived elements in various body fluids, presents a valid alternative to tissue re-biopsy. The liquid biomarkers consisted of circulating free DNA (cfDNA), circulating tumor cells (CTCS), exosomes, and tumor-educated platelets61. These components can be easily isolated from almost a wide range of body fluids, including blood, urine, pleural effusion, and ascites62. Liquid biopsy offers convenient, non-invasive, and the ability to be performed at multiple time-points. Furthermore, it enables the identification of dynamic changes in gene expression within the tumor and the capturing intrinsic tumor heterogeneity63. As a result, liquid biopsy plays an increasingly important role in establishing a diagnosis, detecting molecular characterization, and monitoring mechanisms of resistance in patients with lung cancer64,65,66. The specificity of liquid biopsy for detecting T790M mutation during treatment with first- and second-generation TKIs has also been confirmed65. However, there are cases where the results from tissue and cfDNA genotyping do not match, probably due to technological differences or sampling different tumor cell populations67,68,69. Therefore, patients with T790M-negative plasma results still need a tumor biopsy to identify T790M mutation. Consequently, tissue biopsy and blood-based analyses may have complementary roles in evaluating the genetic alterations for these patients60,68. In this context, radiomics may offer additional information to assist in the implementation of optimal treatment strategies. Cucchiara et al. developed a mode that integrates liquid biopsy and radiomics, demonstrating good performance in identifying the presence of T790M70.
ML is a subset of artificial intelligence (AI) that involves the development of algorithmic models to identify patterns and relationships in data71. The main aim of ML techniques is to create models that can be applied to perform tasks, such as classification, prediction, or estimation. Our retrospective study revealed the feasibility of using 7 ML approaches to predict EGFR T790M mutations after treatment with a first- or second-generation EGFR-TKI. Among these classifiers, RF classification method exhibited the highest performance, with an AUC of 0.87. This finding accords with a study by Saini R et al.72, which reported that RF-based radiomics classifier achieved the best performance (AUC = 0.776) in predicting the Ki-67 expression level in NSCLC. Parmar et al.73 also found that RF classification method showed highest prognostic performance in predicting 2-year patient survival in patients with NSCLC. In addition, RF-based models had best performance in identifying new potent EGFR inhibitors against the resistant T790M mutant74. RF is considered a favorable ML method due to its simple structure, ability to deal with both regression and classification issues, and higher efficiency than compared to methods75. In our study, the RF-based model which integrated the radiomics signatures and the clinical factors achieved an AUC of 0.87. In terms of interpretability, the predictions made by the RF model can be meaningfully explained from a biological standpoint as well76. Additionally, the robustness of the features used in the current study was ensured through the utilization of five-fold cross-validation.
Several limitations of this study should be noted. Firstly, the retrospective nature of the analysis may inevitably result in patient selection bias. Another limitation is the relatively small sample size of patients harboring T790M mutation, which may limit the power of these analyses. However, although larger data sets are more likely to have higher statistical power, radiomics analyses can still be performed with as few as 100 patients77. Further studies with large sample size should be conducted to assess the clinical applications of our models. Thirdly, because all of subjects who were involved in this study were Chinese, generalization of these results should not be made beyond this population. Further studies are required to verify these findings within other racial and ethnic population. Finally, manual segmentation of ROI is both tedious and time consuming, and there may be significant variability among different observers. However, manual segmentation can still be a simple and reliable method, and its reproducibility can be evaluated by interobserver reproducibility analysis78.
In conclusion, this study demonstrates the feasibility of non-invasively predicting EGFR T790M mutation at diagnosis in NSCLC patients following treatment with a first- or second-generation EGFR-TKI using a ML model integrating radiomic features and clinical characteristics. Our results are promising and warrant validation in a larger sample size. These findings indicate that utilizing this method to detect EGFR-T790M mutation could potentially facilitate the selection of accurate and personalized treatment strategies for patients with NSCLC.
Data availability
The original data supporting the conclusions of this paper will be provided unreservedly by the authors to any qualified researcher. The datasets used and/or analyzed during the current study are available from the corresponding author.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Herbst, R. S., Heymach, J. V. & Lippman, S. M. Lung cancer. N. Engl. J. Med. 359, 1367–1380. https://doi.org/10.1056/NEJMra0802714 (2008).
Herbst, R. S., Morgensztern, D. & Boshoff, C. The biology and management of non-small cell lung cancer. Nature 553, 446–454. https://doi.org/10.1038/nature25183 (2018).
Gelatti, A. C. Z., Drilon, A. & Santini, F. C. Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC). Lung Cancer 137, 113–122. https://doi.org/10.1016/j.lungcan.2019.09.017 (2019).
Han, B. et al. EGFR mutation prevalence in Asia-Pacific and Russian patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histology: The IGNITE study. Lung Cancer 113, 37–44. https://doi.org/10.1016/j.lungcan.2017.08.021 (2017).
Mok, T. S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361, 947–957. https://doi.org/10.1056/NEJMoa0810699 (2009).
Wu, Y. L. et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): A randomised, open-label, phase 3 trial. Lancet. Oncol. 18, 1454–1466. https://doi.org/10.1016/s1470-2045(17)30608-3 (2017).
Wu, Y. L. et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet Oncol. 15, 213–222. https://doi.org/10.1016/S1470-2045(13)70604-1 (2014).
Wu, Y. L. et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: Analyses from the phase III, randomized, open-label, ENSURE study. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 26, 1883–1889. https://doi.org/10.1093/annonc/mdv270 (2015).
Koo, D. H. et al. EGFR-TKI is effective regardless of treatment timing in pulmonary adenocarcinoma with EGFR mutation. Cancer Chemother. Pharmacol. 75, 197–206. https://doi.org/10.1007/s00280-014-2631-5 (2015).
Lim, S. M., Syn, N. L., Cho, B. C. & Soo, R. A. Acquired resistance to EGFR targeted therapy in non-small cell lung cancer: Mechanisms and therapeutic strategies. Cancer Treat. Rev. 65, 1–10. https://doi.org/10.1016/j.ctrv.2018.02.006 (2018).
Yun, C. H. et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. U. S. A. 105, 2070–2075. https://doi.org/10.1073/pnas.0709662105 (2008).
Mok, T. S. et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N. Engl. J. Med. 376, 629–640. https://doi.org/10.1056/NEJMoa1612674 (2017).
Soria, J. C. et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 378, 113–125. https://doi.org/10.1056/NEJMoa1713137 (2018).
Takeda, M. & Nakagawa, K. First- and second-generation EGFR-TKIs are all replaced to osimertinib in chemo-naive EGFR mutation-positive non-small cell lung cancer?. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20010146 (2019).
Shah, R. & Lester, J. F. Tyrosine kinase inhibitors for the treatment of EGFR mutation-positive non-small-cell lung cancer: A clash of the generations. Clin. Lung Cancer 21, e216–e228. https://doi.org/10.1016/j.cllc.2019.12.003 (2020).
Hochmair, M. J. et al. Sequential afatinib and osimertinib in patients with EGFR mutation-positive non-small-cell lung cancer: Final analysis of the GioTag study. Future Oncol. 16, 2799–2808. https://doi.org/10.2217/fon-2020-0740 (2020).
Ramalingam, S. S. et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 382, 41–50. https://doi.org/10.1056/NEJMoa1913662 (2020).
Mayerhoefer, M. E. et al. Introduction to radiomics. J. Nucl. Med. 61, 488–495. https://doi.org/10.2967/jnumed.118.222893 (2020).
Thawani, R. et al. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 115, 34–41. https://doi.org/10.1016/j.lungcan.2017.10.015 (2018).
Binczyk, F., Prazuch, W., Bozek, P. & Polanska, J. Radiomics and artificial intelligence in lung cancer screening. Transl. Lung Cancer Res. 10, 1186–1199. https://doi.org/10.21037/tlcr-20-708 (2021).
Avanzo, M., Stancanello, J., Pirrone, G. & Sartor, G. Radiomics and deep learning in lung cancer. Strahlenther. Onkol. 196, 879–887. https://doi.org/10.1007/s00066-020-01625-9 (2020).
Kawahara, D. et al. Prediction of radiation pneumonitis after definitive radiotherapy for locally advanced non-small cell lung cancer using multi-region radiomics analysis. Sci. Rep. 11, 16232. https://doi.org/10.1038/s41598-021-95643-x (2021).
Rossi, G. et al. Radiomic detection of EGFR mutations in NSCLC. Cancer Res. 81, 724–731. https://doi.org/10.1158/0008-5472.CAN-20-0999 (2021).
Wen, Q., Yang, Z., Dai, H., Feng, A. & Li, Q. Radiomics study for predicting the expression of PD-L1 and Tumor mutation burden in non-small cell lung cancer based on CT images and clinicopathological features. Front. Oncol. 11, 620246. https://doi.org/10.3389/fonc.2021.620246 (2021).
Chetan, M. R. & Gleeson, F. V. Radiomics in predicting treatment response in non-small-cell lung cancer: Current status, challenges and future perspectives. Eur. Radiol. 31, 1049–1058. https://doi.org/10.1007/s00330-020-07141-9 (2021).
Lu, J. et al. Machine learning-based radiomics for prediction of epidermal growth factor receptor mutations in lung adenocarcinoma. Dis. Markers 2022, 2056837. https://doi.org/10.1155/2022/2056837 (2022).
Yang, X. et al. Can CT radiomics detect acquired T790M mutation and predict prognosis in advanced lung adenocarcinoma with progression after first- or second-generation EGFR TKIs?. Front. Oncol. 12, 904983. https://doi.org/10.3389/fonc.2022.904983 (2022).
Detterbeck, F. C., Boffa, D. J., Kim, A. W. & Tanoue, L. T. The eighth edition lung cancer stage classification. Chest 151, 193–203. https://doi.org/10.1016/j.chest.2016.10.010 (2017).
Zhou, J. et al. Re-biopsy and liquid biopsy for patients with non-small cell lung cancer after EGFR-tyrosine kinase inhibitor failure. Thorac. Cancer 10, 957–965. https://doi.org/10.1111/1759-7714.13035 (2019).
Zhang, Y. et al. Next-generation sequencing of tissue and circulating tumor DNA: Resistance mechanisms to EGFR targeted therapy in a cohort of patients with advanced non-small cell lung cancer. Cancer Med. 10, 4697–4709. https://doi.org/10.1002/cam4.3948 (2021).
She, Y. et al. The predictive value of CT-based radiomics in differentiating indolent from invasive lung adenocarcinoma in patients with pulmonary nodules. Eur. Radiol. 28, 5121–5128. https://doi.org/10.1007/s00330-018-5509-9 (2018).
Hong, D., Xu, K., Zhang, L., Wan, X. & Guo, Y. Radiomics signature as a predictive factor for EGFR mutations in advanced lung adenocarcinoma. Front. Oncol. 10, 28. https://doi.org/10.3389/fonc.2020.00028 (2020).
Yushkevich, P. A. & Gerig, G. ITK-SNAP: An intractive medical image segmentation tool to meet the need for expert-guided segmentation of complex medical images. IEEE Pulse 8, 54–57. https://doi.org/10.1109/MPUL.2017.2701493 (2017).
Yushkevich, P. A. et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 31, 1116–1128. https://doi.org/10.1016/j.neuroimage.2006.01.015 (2006).
Leijenaar, R. T. et al. Stability of FDG-PET Radiomics features: An integrated analysis of test-retest and inter-observer variability. Acta Oncol. 52, 1391–1397. https://doi.org/10.3109/0284186X.2013.812798 (2013).
Koo, T. K. & Li, M. Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 15, 155–163. https://doi.org/10.1016/j.jcm.2016.02.012 (2016).
Zwanenburg, A. et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295, 328–338. https://doi.org/10.1148/radiol.2020191145 (2020).
Ren, M. et al. MRI-based radiomics analysis for predicting the EGFR mutation based on thoracic spinal metastases in lung adenocarcinoma patients. Med. Phys. 48, 5142–5151. https://doi.org/10.1002/mp.15137 (2021).
Morgado, J. et al. Machine learning and feature selection methods for EGFR mutation status prediction in lung cancer. Appl. Sci. Basel 11, 3273 (2021).
Wilcoxin, F. Probability tables for individual comparisons by ranking methods. Biometrics 3, 119–122 (1947).
Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 16, 385–395. https://doi.org/10.1002/(sici)1097-0258(19970228)16:4%3c385::aid-sim380%3e3.0.co;2-3 (1997).
Li, Z. & Sillanpaa, M. J. Overview of LASSO-related penalized regression methods for quantitative trait mapping and genomic selection. Theor. Appl. Genet. 125, 419–435. https://doi.org/10.1007/s00122-012-1892-9 (2012).
Zhang, Z. Variable selection with stepwise and best subset approaches. Ann. Transl. Med. 4, 136. https://doi.org/10.21037/atm.2016.03.35 (2016).
Abraham, A. et al. Machine learning for neuroimaging with scikit-learn. Front. Neuroinform. 8, 14. https://doi.org/10.3389/fninf.2014.00014 (2014).
Chapman, A. M., Sun, K. Y., Ruestow, P., Cowan, D. M. & Madl, A. K. Lung cancer mutation profile of EGFR, ALK, and KRAS: Meta-analysis and comparison of never and ever smokers. Lung Cancer 102, 122–134. https://doi.org/10.1016/j.lungcan.2016.10.010 (2016).
Wang, S. et al. Value of serum tumor markers for predicting EGFR mutations and positive ALK expression in 1089 Chinese non-small-cell lung cancer patients: A retrospective analysis. Eur. J. Cancer 124, 1–14. https://doi.org/10.1016/j.ejca.2019.10.005 (2020).
Balachandran, V. P., Gonen, M., Smith, J. J. & DeMatteo, R. P. Nomograms in oncology: More than meets the eye. Lancet Oncol. 16, e173-180. https://doi.org/10.1016/S1470-2045(14)71116-7 (2015).
Liu, Z. et al. The applications of radiomics in precision diagnosis and treatment of oncology: Opportunities and challenges. Theranostics 9, 1303–1322. https://doi.org/10.7150/thno.30309 (2019).
Kumar, V. et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 30, 1234–1248. https://doi.org/10.1016/j.mri.2012.06.010 (2012).
Shi, L. et al. Radiomics for response and outcome assessment for non-small cell lung cancer. Technol. Cancer Res. Treat. 17, 1533033818782788. https://doi.org/10.1177/1533033818782788 (2018).
Zhou, M. et al. Non-small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology 286, 307–315. https://doi.org/10.1148/radiol.2017161845 (2018).
Kim, H. et al. Repeat biopsy of patients with acquired resistance to EGFR TKIs: Implications of biopsy-related factors on T790M mutation detection. Eur. Radiol. 28, 861–868. https://doi.org/10.1007/s00330-017-5006-6 (2018).
Hou, D. et al. Different clinicopathologic and computed tomography imaging characteristics of primary and acquired EGFR T790M mutations in patients with non-small-cell lung cancer. Cancer Manag. Res. 13, 6389–6401. https://doi.org/10.2147/CMAR.S323972 (2021).
Koo, H. J. et al. Non-small cell lung cancer with resistance to EGFR-TKI therapy: CT characteristics of T790M mutation-positive cancer. Radiology 289, 227–237. https://doi.org/10.1148/radiol.2018180070 (2018).
Yoshida, T. et al. Standardized uptake value on (18)F-FDG-PET/CT is a predictor of EGFR T790M mutation status in patients with acquired resistance to EGFR-TKIs. Lung Cancer 100, 14–19. https://doi.org/10.1016/j.lungcan.2016.07.022 (2016).
Tang, X. et al. Machine learning-based CT radiomics analysis for prognostic prediction in metastatic non-small cell lung cancer patients with EGFR-T790M mutation receiving third-generation EGFR-TKI osimertinib treatment. Front. Oncol. 11, 719919. https://doi.org/10.3389/fonc.2021.719919 (2021).
Kawamura, T. et al. Clinical factors predicting detection of T790M mutation in rebiopsy for EGFR-mutant non-small-cell lung cancer. Clin. Lung Cancer 19, e247–e252. https://doi.org/10.1016/j.cllc.2017.07.002 (2018).
Oya, Y. et al. Association between EGFR T790M status and progression patterns during initial EGFR-TKI treatment in patients harboring EGFR mutation. Clin. Lung Cancer 18, 698–705. https://doi.org/10.1016/j.cllc.2017.05.004 (2017).
Dal Maso, A. et al. Clinical features and progression pattern of acquired T790M-positive compared With T790M-negative EGFR mutant non-small-cell lung cancer: Catching tumor and clinical heterogeneity over time through liquid biopsy. Clin. Lung Cancer 21, 1–14. https://doi.org/10.1016/j.cllc.2019.07.009 (2020).
Perez-Callejo, D., Romero, A., Provencio, M. & Torrente, M. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl. Lung Cancer Res. 5, 455–465. https://doi.org/10.21037/tlcr.2016.10.07 (2016).
Rolfo, C. et al. Liquid biopsies in lung cancer: The new ambrosia of researchers. Biochim. Biophys. Acta 539–546, 2014. https://doi.org/10.1016/j.bbcan.2014.10.001 (1846).
Del Re, M. et al. Understanding the mechanisms of resistance in EGFR-positive NSCLC: From tissue to liquid biopsy to guide treatment strategy. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20163951 (2019).
Cecchini, M. J. & Yi, E. S. Liquid biopsy is a valuable tool in the diagnosis and management of lung cancer. J. Thorac. Dis. 12, 7048–7056. https://doi.org/10.21037/jtd.2020.04.20 (2020).
Minari, R. et al. Detection of EGFR-activating and T790M mutations using liquid biopsy in patients With EGFR-mutated Non-small-cell lung cancer whose disease has progressed during treatment with first- and second-generation tyrosine kinase inhibitors: A multicenter real-life retrospective study. Clin. Lung Cancer 21, e464–e473. https://doi.org/10.1016/j.cllc.2020.02.021 (2020).
Buder, A. et al. Cell-free plasma DNA-guided treatment with osimertinib in patients with advanced EGFR-mutated NSCLC. J. Thorac. Oncol. 13, 821–830. https://doi.org/10.1016/j.jtho.2018.02.014 (2018).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. J. Clin. Oncol. 36, 1631–1641. https://doi.org/10.1200/JCO.2017.76.8671 (2018).
Sundaresan, T. K. et al. Detection of T790M, the acquired resistance EGFR Mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin. Cancer Res. 22, 1103–1110. https://doi.org/10.1158/1078-0432.CCR-15-1031 (2016).
Oxnard, G. R. et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J. Clin. Oncol. 34, 3375–3382. https://doi.org/10.1200/JCO.2016.66.7162 (2016).
Cucchiara, F. et al. Integrating liquid biopsy and radiomics to monitor clonal heterogeneity of EGFR-positive non-small cell lung cancer. Front. Oncol. 10, 593831. https://doi.org/10.3389/fonc.2020.593831 (2020).
Kourou, K., Exarchos, T. P., Exarchos, K. P., Karamouzis, M. V. & Fotiadis, D. I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 13, 8–17. https://doi.org/10.1016/j.csbj.2014.11.005 (2015).
Gu, Q. et al. Machine learning-based radiomics strategy for prediction of cell proliferation in non-small cell lung cancer. Eur. J. Radiol. 118, 32–37. https://doi.org/10.1016/j.ejrad.2019.06.025 (2019).
Parmar, C., Grossmann, P., Bussink, J., Lambin, P. & Aerts, H. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 5, 13087. https://doi.org/10.1038/srep13087 (2015).
Saini, R., Fatima, S. & Agarwal, S. M. TMLRpred: A machine learning classification model to distinguish reversible EGFR double mutant inhibitors. Chem. Biol. Drug Des. 96, 921–930. https://doi.org/10.1111/cbdd.13697 (2020).
Savargiv, M., Masoumi, B. & Keyvanpour, M. R. A new random forest algorithm based on learning automata. Comput. Intell. Neurosci. 2021, 5572781. https://doi.org/10.1155/2021/5572781 (2021).
Marchese Robinson, R. L. et al. Comparison of the predictive performance and interpretability of random forest and linear models on benchmark data sets. J. Chem. Inf. Model. 28, 1773–1792. https://doi.org/10.1021/acs.jcim.6b00753 (2017).
Gillies, R. J., Kinahan, P. E. & Hricak, H. Radiomics: images are more than pictures, they are data. Radiology 278, 563–577. https://doi.org/10.1148/radiol.2015151169 (2016).
Lohmann, P., Bousabarah, K., Hoevels, M. & Treuer, H. Radiomics in radiation oncology-basics, methods, and limitations. Strahlenther. Onkol. 196, 848–855. https://doi.org/10.1007/s00066-020-01663-3 (2020).
Funding
This work was supported by the Collaborative Innovation Center for Intelligent Molecules with Multi-effects and Nanomedicine (Grant No. 2019–01), Shandong province, China. The funders did not play a role in design and conduct of study, collection and analysis of data, preparation, review, or approval of the manuscript, and decision of submitting the manuscript for publication.
Author information
Authors and Affiliations
Contributions
Conception and design: J.L., X.J., D.L., Y.J. Collection and assembly of data: J.L., X.J., Y.J., P.F., X.L., S.Y., Z.G., A.Z. Data analysis and interpretation: J.L., X.J., X.L., G.L., W.L., P.F., Y.J., D.L. Manuscript writing: All authors, Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lu, J., Ji, X., Liu, X. et al. Machine learning-based radiomics strategy for prediction of acquired EGFR T790M mutation following treatment with EGFR-TKI in NSCLC. Sci Rep 14, 446 (2024). https://doi.org/10.1038/s41598-023-50984-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-50984-7
- Springer Nature Limited