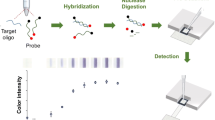
Abstract
A novel integrated detection system that introduces a paper-chip-based molecular detection strategy into a polydimethylsiloxane (PDMS) microchip and temperature control system was developed for on-site colorimetric detection of DNA. For the paper chip-based detection strategy, a padlock probe DNA (PLP)-mediated rolling circle amplification (RCA) reaction for signal amplification and a radial flow assay according to the Au-probe labeling strategy for visualization were optimized and applied for DNA detection. In the PDMS chip, the reactions for ligation of target-dependent PLP, RCA, and labeling were performed one-step under isothermal temperature in a single chamber, and one drop of the final reaction solution was loaded onto the paper chip to form a radial colorimetric signal. To create an optimal analysis environment, not only the optimization of molecular reactions for DNA detection but also the chamber shape of the PDMS chip and temperature control system were successfully verified. Our results indicate that a detection limit of 14.7 nM of DNA was achieved, and non-specific DNAs with a single-base mismatch at the target DNA were selectively discriminated. This integrated detection system can be applied not only for single nucleotide polymorphism identification, but also for pathogen gene detection. The adoption of inexpensive paper and PDMS chips allows the fabrication of cost-effective detection systems. Moreover, it is very suitable for operation in various resource-limited locations by adopting a highly portable and user-friendly detection method that minimizes the use of large and expensive equipment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Sensitive nucleic acid detection is crucial in disease diagnosis, gene therapy, forensic and biomedical research, and food safety [1,2,3]. Particularly, with the recent COVID-19 pandemic, DNA detection technology is drawing attention in fields that require investigation of viruses and bacteria [4, 5]. The standard method for laboratory DNA detection is polymerase chain reaction (PCR), which selectively amplifies a desired gene site using a DNA polymerase and a pair of target-specific primers with high thermal stability, high sensitivity, and reliability [6,7,8]. However, PCR requires large and expensive thermal cyclers and optical analysis equipment, along with skilled personnel because detailed experimental designs and controls are required to obtain satisfactory results [9, 10]. Hence, the application of PCR for DNA detection in a resource-limited environment is difficult, thereby limiting its application to laboratories with appropriate facilities and personnel. To overcome the limitations of conventional PCR-based detection methods, various biosensors to which gene isothermal amplification technology is applied have been developed [11].
Representative gene isothermal amplification methods include nucleic acid sequence-based amplification [12], loop-mediated isothermal amplification [13,14,15], recombinase polymerase amplification [16], strand displacement amplification [17], and rolling circle amplification (RCA) [18]. RCA is a method of amplifying tandemly linked copies of a complementary sequence of a circular template using circularizable oligonucleotides (for example, padlock probe DNA) and linear single-stranded DNA (for example, primers) capable of annealing [19, 20]. Direct binding and stringent ligation of the padlock probe DNA (PLP) toward the target DNA is useful for the selective detection of gene sequences and variants, and displacement of the synthesized long DNA strand during replication can be used to bind the sensing probes to generate an amplified signal [21, 22].
Typically, an isothermal amplification method, such as RCA, does not include a complicated temperature control step; therefore, it has the advantage of avoiding the use of more complex equipment than the PCR-based detection method, and the analysis procedure is simple [23,24,25]. Furthermore, research is being actively conducted to integrate biosensors into a detection platform with excellent portability and economy in various applications requiring on-site detection for the diagnosis of diseases or rapid detection of hazardous substances [26,27,28,29,30]. Among them, paper has been successfully used as a platform for various types of point-of-care testing (POCT), such as lateral flow assays [31], microfluidic paper-based analytical devices [32], and dipstick assays [33] based on advantages, such as high portability, low cost, and easy operation [34]. Polydimethylsiloxane (PDMS) is an elastomer with excellent biocompatibility, mechanical properties, high optical transparency, and chemical stability [35, 36]. Particularly, the ability to easily fabricate a desired size and shape has led to the use of PDMS in microelectromechanical and microfluidic systems for POCT [37,38,39]. The visual analysis strategy suitable for POCT does not require expensive equipment for signal analysis, and the results can be analyzed intuitively, so it can be introduced into a portable detection chip to ease on-site analysis. Several researches have shown that additional strategies are required to regulate complex external environmental factors, including natural light, for consistent and accurate colorimetric signal processing [40, 41].
In this study, we demonstrate a novel integrated detection system by introducing a paper-chip-based molecular detection strategy for the colorimetric detection of DNA into a PDMS microchip and temperature control system. For paper-chip-based DNA detection, a PLP-mediated RCA reaction for signal amplification and a radial flow assay according to the Au-probe labeling strategy for visualization were applied. These molecular reactions for DNA detection occur in one-step at uniform temperature and can be successfully implemented in a portable PDMS microchip-based detection platform, providing user-friendly and easy operation. In addition, colorimetric images of the paper-chips were acquired using a conventional digital scanner to achieve consistent colorimetric signal analysis without interference from complex external environments.
2 Results and Discussion
2.1 Design and Fabrication of Microchip-Based Detection Platform
The microchip was manufactured by pouring and curing polydimethylsiloxane (PDMS) prepolymer into a master mold manufactured using high-resolution stereolithography (SLA) 3D printing (Fig. 1A). The microchip comprises two PDMS layers, and is manufactured at 11.5 × 3.9 cm dimensions. The length from the inlet to the outlet was 2.6 cm, the microchannel width was 0.6 mm, and the chamber was manufactured to a depth of 1 mm and a volume of 68.3 ± 1.3 μL. The RCA microchip was fabricated using PDMS to have the flexibility to facilitate the release of the final products after the reaction. After the RCA reaction, 23.88 ± 1.73 μL of the product (1 drop) was released through the outlet of the chip when pressure was manually applied vertically to the chamber (Table S1, Supplementary Material).
Design and fabrication of the microchip and temperature control system. A Schematic diagram (left) and an optical image (right) of the RCA microchip. B Schematic diagram of the heater of the temperature control system. C Optical image of the temperature control system consisting of a power supplier, RCA microchip, heater, and temperature controller. Scale bar is 5 cm
Power was supplied to the temperature controller and heater through a 12 V DC power supplier (Fig. 1B). Three ceramic heaters, each measuring 5 × 5 cm in size, were connected to cover all the microchips (Fig. 1C). A copper plate with a high thermal conductivity (15 × 4.5 cm) was placed on it to transfer the heat of the heater to the microchip with minimal loss. The upper microchip case was printed with a size of 15 × 8 × 4 mm using an FDM (Fused Deposition Modeling; M16, Moment, Seoul, Korea) 3D printer. This case was designed to have a space in the middle, such that the microchip was placed in a fixed position on the heater and fabricated to package the entire heater.
2.2 Description of Paper-Based Colorimetric Detection Strategy
The detailed process of the paper-based colorimetric detection strategy is illustrated in Fig. 2. A padlock probe designed for target DNA detection forms a partial double helix complex containing a nick when in the presence of the target DNA. The nick is linked by a catalytic reaction of T4 ligase to produce single-stranded circular DNA, which is used as a template for the RCA reaction. Furthermore, because the target DNA hybridized with the circular DNA can serve as a primer for the RCA reaction by providing a 3-terminal OH group, additional primers are not required for the RCA reaction. The RCA reaction catalyzed by Phi29 DNA polymerase under isothermal conditions produces a highly repetitive long single stranded DNA (ssDNA) sequence complementary to the PLP sequence, which is hybridized with a gold nanoparticle-conjugated DNA probe (Au-probe) to form a large complex. Thus, the series of reactions of ligation, RCA, and labeling in the microchip chamber are performed in one-step under 30 °C incubation condition. The final reaction solution in the microchip chamber was loaded with one drop on the nitrocellulose paper chip (Fig. S1, Supplementary Material) by hand pressure and dried to form a red circular colorimetric pattern (see Supplementary Material, Video Clip). The paper chip is imaged using a scanner and converted into an 8-bit black and white image; the colorimetric intensity of the circular area (diameter 6 mm, 70 × 70 pixels) in the center of the colorimetric pattern is quantified “gray value” representing the brightness of pixels using ImageJ software (version 1.53e). The circular area for signal analysis included all the inner spot areas formed when the reaction solution was dropped, and the gray value of the spot increased as the concentration of the target DNA increased. It is a phenomenon in which the diffusion distance in the paper chip is shortened because the Au-probes hybridize to the RCA product to form a large complex [22]. As a result, the color intensities of the RCA reaction solutions with different target DNA concentrations in the PDMS chip were indistinguishable with the naked eye, but concentric spots of different patterns that could be visually recognized after introducing the solution into the paper chip.
2.3 Optimizing Condition of the Temperature Controller
When the temperature of the temperature controller was increased from 25 to 80 °C over 5 °C intervals, the chamber temperature was measured (Fig. 3A). As the input temperature of the controller increased, the output temperature of the chamber increased linearly, showing a relationship of y = 0.86x + 4.98 (R2 = 0.996). Therefore, we confirmed that the chamber temperature can be sufficiently controlled using a temperature controller.
Temperature optimization of the PDMS microchip by the temperature control system. A The measured output temperature of the microchip chamber according to the input temperature controlled by the temperature controller. B Temperature maintenance performance test in the microchip chamber during one-step isothermal reaction for ligation and RCA
To determine whether the temperature was transmitted evenly to the entire microchip, the temperatures of the eight chambers were measured and compared under the same temperature conditions (Table 1). The microchamber temperature was designated as 30 °C, which is the isothermal amplification condition for RCA, and the controller temperature was measured. A minute temperature difference was measured between the chambers, and the temperature difference increased toward the edge. However, the temperature of the eight chambers did not show statistically significant variation (one-way analysis of variance [ANOVA], p-value = 0.56 > 0.05). When the controller temperature averaged 30.5 ± 0.4 °C, the chamber was maintained at 30 °C. Therefore, it was decided to input 30.5 °C to the temperature controller for on-chip RCA.
The one-step isothermal reaction lasted 80 min at 30 °C, when probe ligation and RCA proceeded, and 10 min at 65 °C, when enzyme inactivation progressed. During the entire reaction time, we checked whether the temperature system operated stably, and the temperature was maintained uniformly at 5 min intervals (Fig. 3B). In the 30 °C setting section, the chamber temperature was stably maintained within the range of 29.8–30.5 °C. When the temperature increased from the 30 to 65 °C section, it took 5 min to stabilize the temperature change. Furthermore, in the 65 °C setting section, the chamber temperature was maintained at 64.8–65.6 °C. Therefore, it was verified that an accurate and rapid RCA reaction is possible in the microchip by optimizing the temperature conditions of the controller and microchamber.
2.4 Padlock Probe Verification and Optimization of One-Step Enzymatic Reaction Parameters
Circularization of the padlock probe designed for target DNA detection was verified. First, as depicted in the results of urea-polyacrylamide gel electrophoresis (PAGE), linear PLP was observed as a single band corresponding to the 40 bp DNA ladder, whereas PLP self-ligated by CircLigase catalysis was observed as a single band corresponding to the 60 bp DNA ladder (Fig. S2A, Supplementary Material). This result is consistent with our previous study showing that circularized ssDNA migrates later than linear ssDNA in urea-PAGE analysis [22]. Target DNA-dependent circularization of PLP by the catalytic reaction of T4 ligase was confirmed. To this end, a ligation reaction, according to the PLP concentration, was performed in the presence of 1 µM target DNA. As illustrated in the urea-PAGE image (Fig. S2B, Supplementary Material), it was confirmed that the band of approximately 60 bp corresponding to circularized PLP gradually thickened as the amount of PLP increased. This result shows that linear PLP can be circularized in a target DNA-dependent manner using T4 ligase catalysis.
The optimal conditions for one-step enzymatic and colorimetric reactions were explored for the efficient implementation of a paper-based colorimetric detection strategy. First, the appropriate concentration of the Au-probe was investigated for the optimization of colorimetric signal formation on a paper chip. The Au-probes were prepared in a slightly modified manner following a previous study [22], introducing biotin-labeled DNA into streptavidin-coated gold nanoparticles, and the target DNA of 0 or 300 nM was analyzed in triplicates under conditions of various Au-probe concentrations (1, 1.5, 2, and 2.5 optical densities [OD]). The experimental results showed that the Δ gray value increased as the concentration of the Au-probe increased, which then saturated at a condition of 2 OD (Fig. 4A). The Δ gray value was determined by subtracting the gray value obtained in the absence of target DNA from the gray value obtained in the presence of 300 nM of target DNA. Measured mean Δ gray values were as follows: 55.38 ± 4.96, 62.83 ± 6.13, 69.38 ± 3.88, and 67.55 ± 2.65, in the 1, 1.5, 2, and 2.5 OD under Au-probe conditions, respectively. Therefore, 2 OD with the highest Δ gray value was determined as the optimal Au-probe concentration.
Optimization of enzymatic reaction parameters. A Au-probe concentration. B Incubation conditions. Condition A: 30 °C for 40 min for integration of ligation and RCA, condition B: 22 °C for 10 min for ligation and 30 °C for 30 min for RCA, condition C: 22 °C for 20 min for ligation and 30 °C for 20 min for RCA
For easy and convenient biosensor development, simplification of the analysis steps is important because complex and multistep enzymatic reactions are one of the factors that complicate the detection procedure and increase the time required. Therefore, we integrated the ligation, RCA, and labeling reactions into a single step to streamline the analytical procedure of the proposed detection method. The optimal incubation temperature for the enzymatic reaction recommended by each manufacturer was 22 °C for T4 ligase and 30 °C for Phi29 DNA polymerase. To integrate both reactions, conditions with a combination of incubation temperatures of 22 °C or 30 °C and various incubation times were investigated in triplicates under 0 or 300 nM of target DNA; the results are shown in Fig. 4B. For a total incubation time of 40 min, there was no significant difference in Δ gray value results under the three different conditions of maintaining a single temperature of 30 °C for 40 min to integrate the ligation and RCA (condition A), or separating the ligation step at 22 °C for 10 min (condition B) or 20 min (condition C) from RCA step. Measured mean Δ gray values were as follows: 70.01 ± 5.83, 69.55 ± 6.13, and 65.30 ± 6.91, in condition A, B, and C, respectively. In the graph depicting the relationship between the incubation condition and mean Δ gray value (Fig. 4B), the Δ gray value was the highest at 70.01 under condition A. These results indicate that the target DNA-dependent ligation reaction of PLP works well, even at a single temperature condition of 30 °C. Moreover, by integrating the two-step enzymatic reaction into a single step, the complicated temperature conversion process was omitted and intact isothermal amplification conditions were established.
2.5 Selectivity Test of the Proposed Assay
To confirm the selectivity of the proposed detection method for target DNA, three types of mismatched target DNA with a single base mismatch (MT1, MT2, and MT3), and the target DNA, were tested using a microchip-based detection platform. As a negative control, a 63-mer length of control PLP (C-PLP) with no complementary site to the target DNA was prepared and tested using the target DNA. Furthermore, samples mixed with MT DNAs and the target DNA were tested to verify cross-reactivity among the various DNA samples. All samples were tested in triplicates. The results of the experiment confirmed that a coffee-ring-like pattern was generated in the negative control and MT DNA samples, whereas a strong colorimetric signal was generated up to the center of the circular pattern in the target DNA and mixed samples (Fig. 5A). These differences displayed such a contrast that they could be clearly distinguished with the naked eye. The gray values for each sample (negative control, MT1, MT2, MT3, target DNA, and mixture) measured through image analysis are 25.87 ± 3.54, 63.03 ± 11.85, 56.13 ± 5.20, 55.47 ± 3.80, 141.57 ± 16.20, and 146.20 ± 2.65, respectively (Fig. 5B). The target and mixed samples indicated approximately seven times greater gray values than the negative control. In contrast, a slight increase in gray values was observed in comparison to the negative control in MT DNA samples. These results are considered acceptable, given that the MT DNAs showed 95% sequence similarity with the target DNA only by a single base difference.
2.6 Sensitivity and Recovery Test of the Proposed Assay
The detection performance of the proposed method for the quantitative analysis of the target DNA was evaluated. To this end, nine samples with different target DNA concentrations (0, 1, 10, 50, 100, 200, 300, 400, and 500 nM) were tested in triplicates (Fig. 6A); the measured gray values were 26.37 ± 5.20, 64.83 ± 1.42, 72.00 ± 10.49, 88.83 ± 4.37, 114.57 ± 4.17, 140.50 ± 11.23, 160.20 ± 6.90, 163.13 ± 2.63, and 170.50 ± 8.88, respectively. As the concentration of target DNA increased, the spot signal tended to become stronger (Fig. 6B). The measured gray values showed linearity between 1 and 300 nM target DNA concentrations. The linear relationship between the target DNA and the gray value was y = 0.3186x + 71.721, and the R2 value was 0.9648 (Fig. 6C). The detection limit (LOD) for the target DNA was 14.7 nM according to the following formula:
A Gray values of the nine target DNA samples with different concentration (0, 1, 10, 50, 100, 200, 300, 400, and 500 nM) tested by the detection platform. B Digital scanned image of the paper chip after drop-drying of six reaction solutions tested by the detection platform. Scale bar is 15 mm. C Linear correlation between target DNA concentration (1, 10, 50, 100, 200, and 300 nM) and gray values. The error bars represent the standard deviations of the mean values from three individual experiments set (n = 3)
In this system, increasing the RCA reaction time (DNA amplification time) improves sensitivity but reduces the rapidity of field detection. Therefore, we focused our efforts on integrating the biosensor into a portable platform for easy operation in the field where equipment is limited rather than improving the performance of the biosensor.
To evaluate the accuracy and reproducibility of this detection method, target DNA samples of arbitrary concentrations (80, 180, and 250 nM) within the concentration range of the calibration curve were analyzed, and the coefficient of variation and recovery rate were evaluated. The results are listed in Table 2. The coefficients of variation for the target DNA concentrations of 80, 180, and 250 nM samples were 14.2, 8.0, and 1.6%, respectively, and the recovery rates were 111.6, 105.7, and 99.8, respectively. These results confirm that the reproducibility and precision of this detection method are excellent.
3 Conclusions
In conclusion, a novel integrated detection system was developed by introducing a paper-chip-based molecular detection strategy into a PDMS microchip and a temperature control system for on-site colorimetric detection of DNA. The paper chip-based detection strategy for the molecular detection of DNA was optimized, and two enzymatic and labeling reactions were performed one-step under single temperature conditions to improve the simplicity of the analysis. The PDMS microchip and temperature control system designed in this study were also successfully optimized, providing high economic efficiency, portability, and user convenience to implement the paper-chip-based detection strategy in a resource-limited environment. The selectivity of the integrated detection system was demonstrated using a target DNA sample, a single-base mismatch DNA sample, and their mixture. Furthermore, a detection limit of 14.7 nM was achieved and the accuracy and reproducibility of the assay for DNA detection were evaluated. Although this integrated detection system did not achieve ultra-high sensitivity, it provides many advantages for high user convenience and on-site detection. Therefore, it is a promising platform for POCT applications requiring nucleic acid detection.
4 Materials and Methods
4.1 Chemicals and Reagents
All synthetic DNA oligomers used in this study were obtained from Bioneer (Daejeon, Korea), and are listed in Table S2 (Supplementary Material). Phi29 DNA polymerase (10 U/μL) was purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). T4 DNA ligase (5 U/μL) and dNTP mixture (10 mM) were purchased from Invitrogen (Carlsbad, CA, USA). CircLigase (100 U/μL) was purchased from Epicenter Biotechnologies (Madison, WI, USA). Streptavidin-coated Au nanoparticle solution (20 nm) was purchased from Cytodiagnostics (Burlington, ON, Canada). Urea-polyacrylamide gel (15%) was purchased from Bio-Rad (Hercules, CA, USA). Exonucleases I and III were purchased from New England Biolabs (Ipswich, MA, USA). SYBR Green II was purchased from Life Technologies (Carlsbad, CA, USA). Nitrocellulose membranes (CNPF-SN12, width 25 mm, pore size 10 μm) were purchased from MDI Membrane Technologies (Ambala, India). An upper plastic card (width 25 mm, length 275 mm, 12 holes with a diameter of 15 mm) and a bottom plastic card (width 25 mm, length 300 mm) were purchased from Again BS (Hanam, Korea). Polydimethyl siloxane (PDMS) was purchased from Dow Corning (Midland, TX, USA). The biopsy punch was purchased from Kai Medical (Seoul, Korea). Thermometers and thermocouples were purchased from Omega Engineering (Seongnam, Korea). Thin-film temperature sensors, temperature controllers, power suppliers, and ceramic heaters were purchased from SCIPIA (Gwangju, Korea). Copper and insulation plates were purchased from NAVIMRO (Seoul, Korea).
4.2 PDMS Microchip Fabrication and Optimizing Condition of the Temperature Controller
The PDMS microchip comprised two layers, and was designed to contain eight reaction units (Fig. 1A). Each unit comprised an inlet, a reaction chamber, and an outlet, and the reaction chamber was designed with a streamlined top and side to prevent the formation of bubbles when the solution is injected. An outlet was placed between the layers to facilitate the release of solution after the RCA reaction. The RCA microchip was designed using computer-aided design (CAD) software (3DS Max, USA), and the master mold was manufactured using a stereolithography 3D printer (Form 3, Formlabs Inc., Somerville, MA, USA). The PDMS base and cross-linker were mixed at a ratio of 10:1, poured onto the mold, degassed, and cured at 80 °C for 18–24 h. Then, the PDMS layers were bonded using a plasma device (BD-10A high-frequency generator, Electro-Technic Products, Inc., Chicago, IL, USA).
A temperature control system consisting of a power supplier, temperature controller, and heater was fabricated to perform RCA on the microchip (Fig. 1B). The heater comprised (1) an upper microchip case to determine the microchip position and to minimize heat dissipation, (2) a copper plate to provide uniform heat to the microchip, (3) a ceramic heater to heat the microchip to a specified temperature, (4) a thin-film temperature sensor to accurately detect the temperature of the heater, and (5) an insulating plate to minimize the heat dissipation of the heater (Fig. 1C). This system was designed to set the desired temperature with a temperature controller, measure the temperature with a thin-film temperature sensor between the copper plate and the microchip, and then raise the ceramic heater to the desired temperature. If the temperature rises above the set temperature, the ceramic heater stops working until it drops to the set temperature.
Because the temperature controller controls the temperature based on a thin-film temperature sensor attached to the copper plate, there is a difference from the temperature of the RCA microchip chamber. Therefore, we optimized the temperature conditions by measuring the temperature input values of the temperature controller and microchip chamber. The temperature of the microchip chamber was measured using a thermometer (DP32PT, Omega Engineering) connected to a thermocouple (5TC-TT-K-30-36, Omega Engineering). First, when the set temperature of the controller was increased by 5 °C intervals from 25 to 80 °C, the temperature change in the microchamber was measured. Second, an experiment was conducted to verify the accuracy of multiple reactions by comparing the temperature differences among the eight chambers in a microchip. Third, the microchamber temperature was measured at 5 min intervals to check whether the temperature was stably maintained during the entire gene isothermal amplification process. Temperature measurements was carried out at 30 °C for 80 min (for ligation and RCA) and at 65 °C for 10 min (for inactivation). All experiments were conducted at least thrice. The mean and standard deviation were calculated from the data obtained with at least three replicates. One-way ANOVA was used to determine the statistical significance of comparing temperatures among the chambers in a single microchip. For all data, a p-value < 0.05 was considered statistically significant.
4.3 Optimization of One-Step Enzyme Reaction Parameters
To determine the optimal Au-probe concentration for colorimetric signal formation, four Au-probe concentrations (1, 1.5, 2, and 2.5 OD) were tested in triplicates. To this end, 20 µL of reaction solution (1 × RCA reaction buffer, target DNA, and PLP) in a PCR tube was incubated at 95 °C for 5 min and then slowly cooled to 30 °C to induce the hybridization of the PLP with the target DNA. Subsequently, 1 × RCA reaction buffer, 1 mM dNTPs, 20 units of phi29 DNA pol, 5 units of T4 DNA ligase, and various concentrations of Au-probe were added to the reaction solution to prepare a final 60 µL mixture in a PCR tube. The final concentration of PLP was 500 nM, and that of the target DNA was 0 or 300 nM. This mixture was incubated at 22 °C for 15 min, 30 °C for 30 min, and then inactivated at 65 °C for 10 min. Then, 20 µL of the reaction solution was dropped onto the paper chip using a pipette. After drying for 20 min, the paper chip was imaged using a scanner (SL-M2670N, Samsung, Seoul, Korea; Fig. S3A, Supplementary Material). This image was then converted to an 8-bit black and white image and the gray values of the spot-centered circular area (diameter 6 mm, 70 × 70 pixels) using ImageJ software (National Institutes of Health, Bethesda, MD, USA; version 1.53e, Fig. S3B, Supplementary Material).
Various incubation temperature conditions were investigated in triplicates to effectively induce ligation of the PLP hybridized with the target DNA and RCA reaction. Incubation condition A was set to incubate at 30 °C for 40 min to integrate the ligation and RCA, condition B was set to incubate at 22 °C for 10 min for ligation and 30 °C for 30 min for RCA, and condition C was set to incubate at 22 °C for 20 min for ligation and at 30 °C for 20 min for RCA. Sixty microliters of the total reaction mixture containing Au-probes of 2 OD and target DNA of 0 or 300 nM, identical to the Au-probe concentration optimization experiment above, was incubated individually under A, B, and C conditions, and finally inactivated at 65 °C for 10 min for enzyme inactivation. Then, 20 µL of the reaction solution was dropped onto the paper chip using a pipette and dried for 20 min. The paper chip was imaged and converted to an 8-bit black and white image, and the gray values of the spot-centered circular area were measured using ImageJ software.
4.4 Selectivity Test of the Proposed Method
To confirm the selectivity of this detection method for the target DNA, three types of mismatched target DNAs with a single-base mismatch (MT1, MT2, and MT3) were tested using a microchip-based platform under optimized conditions. C-PLP, which is non-complementary to the target gene, was used as a negative control. First, 20 µL of reaction solution (1 × RCA reaction buffer, target DNA or MTs, and PLP or C-PLP) in a PCR tube was incubated at 95 °C for 5 min and then slowly cooled to 30 °C. Subsequently, 1 × RCA reaction buffer, 1 mM dNTPs, 20 units of Phi29 DNA pol, 5 units of T4 DNA ligase, and 2 OD of Au-probe were added to the reaction solution to prepare 60 µL of the final mixture in a microchip. The final concentration of PLP and C-PLP was 500 nM and that of the target DNA and MTs was 200 nM. The microchip was incubated at 30 °C for 80 min and then inactivated at 65 °C for 10 min using the portable temperature control system fabricated in this study. One drop (approximately 24 µL) of the final reaction solution in the microchip was manually loaded onto the paper chip and dried for 20 min. Imaging and analysis of paper chips were performed as previously described.
4.5 Quantitative Analysis for DNA Samples and Reproducibility Evaluation of the Developed Method
To confirm the trend of gray values according to the concentration of target DNA, various concentrations of target DNA (0, 1, 10, 50, 100, 200, 300, 400, and 500 nM) were individually tested thrice under optimal conditions, including 500 nM PLP. Finally, to evaluate the performance of this detection method, target DNA samples prepared at arbitrary concentrations (80, 180, and 250 nM) within the concentration range of the standard curve were individually tested thrice, and the mean and standard deviation of the quantitative values, coefficients of variation values, and recovery rates were calculated based on this experiment. All experiments described in this section were performed with the same procedures and methods as those in the Sect. 4.4.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Tong, Y.K., Lo, Y.D.: Diagnostic developments involving cell-free (circulating) nucleic acids. Clin. Chim. Acta 363, 187–196 (2006). https://doi.org/10.1016/j.cccn.2005.05.048
Wood, C.S., Thomas, M.R., Budd, J., Mashamba-Thompson, T.P., Herbst, K., Pillay, D., Peeling, R.W., Johnson, A.M., McKendry, R.A., Stevens, M.M.: Taking connected mobile-health diagnostics of infectious diseases to the field. Nature 566, 467–474 (2019). https://doi.org/10.1038/s41586-019-0956-2
McKillip, J.L., Drake, M.: Real-time nucleic acid–based detection methods for pathogenic bacteria in food. J. Food Prot. 67, 823–832 (2004). https://doi.org/10.4315/0362-028X-67.4.823
Yao, L., Zhu, W., Shi, J., Xu, T., Qu, G., Zhou, W., Yu, X.F., Zhang, X., Jiang, G.: Detection of coronavirus in environmental surveillance and risk monitoring for pandemic control. Chem. Soc. Rev. 50, 3656–3676 (2021)
Shen, M., Zhou, Y., Ye, J., Al-Maskri, A.A.A., Kang, Y., Zeng, S., Cai, S.: Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 10, 97–101 (2020). https://doi.org/10.1016/j.jpha.2020.02.010
Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G., Erlich, H.: Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Biotechnol. Ser. (1992). https://doi.org/10.1101/sqb.1986.051.01.032
Yang, S., Rothman, R.E.: PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 4, 337–348 (2004). https://doi.org/10.1016/S1473-3099(04)01044-8
Botes, M., de Kwaadsteniet, M., Cloete, T.E.: Application of quantitative PCR for the detection of microorganisms in water. Anal. Bioanal. Chem. 405, 91–108 (2013). https://doi.org/10.1007/s00216-012-6399-3
Hajia, M.: Limitations of different PCR protocols used in diagnostic laboratories: a short review. Mod. Med. Lab. J. 1, 1–6 (2018) http://modernmedlab.com/article-1-35-en.html
Toze, S.: PCR and the detection of microbial pathogens in water and wastewater. Water Res. 33, 3545–3556 (1999). https://doi.org/10.1016/S0043-1354(99)00071-8
Zhao, Y., Chen, F., Li, Q., Wang, L., Fan, C.: Isothermal amplification of nucleic acids. Chem. Rev. 115, 12491–12545 (2015). https://doi.org/10.1021/acs.chemrev.5b00428
Guoshuai, J., Xiaomeng, X., Zengdan, G., Xingxing, H., Qi, P., Hanbing, Z., Yi, W.: A rapid and high sensitivity RNA detection based on NASBA and G4-ThT fluorescent biosensor. Sci. Rep. 12, 1–9 (2022). https://doi.org/10.1038/s41598-022-14107-y
Kim, J.H., Kang, M., Park, E., Chung, D.R., Kim, J., Hwang, E.S.: A simple and multiplex loop-mediated isothermal amplification (LAMP) assay for rapid detection of SARS-CoV. BioChip J. 13, 341–351 (2019)
Jang, M., Kim, S.: Inhibition of non-specific amplification in loop-mediated isothermal amplification via tetramethylammonium chloride. BioChip J. 16, 326–333 (2022). https://doi.org/10.1007/s13206-022-00070-3
Nguyen, D.V., Nguyen, V.H., Seo, T.S.: Quantification of colorimetric loop-mediated isothermal amplification process. BioChip J. 13, 158–164 (2019). https://doi.org/10.1007/s13206-019-3206-7
Woo, A., Jung, H.S., Kim, D.H., Park, S.G., Lee, M.Y.: Rapid and sensitive multiplex molecular diagnosis of respiratory pathogens using plasmonic isothermal RPA array chip. Biosens. Bioelectron. 182, 113167 (2021). https://doi.org/10.1016/j.bios.2021.113167
Zhou, W., Hu, L., Ying, L., Zhao, Z., Chu, P.K., Yu, X.F.: A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 9, 1–11 (2018). https://doi.org/10.1038/s41467-018-07324-5
Kim, D., Lee, J., Park, S., Park, J., Seo, M.J., Rhee, W.J., Kim, E.: Target-responsive template structure switching-mediated exponential rolling circle amplification for the direct and sensitive detection of microRNA. BioChip J. 16, 422–432 (2022). https://doi.org/10.1007/s13206-022-00071-2
Lee, H.N., Lee, J., Kang, Y.K., Lee, J.H., Yang, S., Chung, H.J.: A lateral flow assay for nucleic acid detection based on rolling circle amplification using capture ligand-modified oligonucleotides. BioChip J. 16, 441–450 (2022). https://doi.org/10.1007/s13206-022-00080-1
Soares, R.R., Varela, J.C., Neogi, U., Ciftci, S., Ashokkumar, M., Pinto, I.F., Nilsson, M., Madaboosi, N., Russom, A.: Sub-attomole detection of HIV-1 using padlock probes and rolling circle amplification combined with microfluidic affinity chromatography. Biosens. Bioelectron. 166, 112442 (2020). https://doi.org/10.1016/j.bios.2020.112442
Tong, P., Zhao, W.W., Zhang, L., Xu, J.J., Chen, H.Y.: Double-probe signal enhancing strategy for toxin aptasensing based on rolling circle amplification. Biosens. Bioelectron. 33, 146–151 (2012). https://doi.org/10.1016/j.bios.2011.12.042
Kim, T.Y., Lim, M.C., Woo, M.A., Jun, B.H.: Radial flow assay using gold nanoparticles and rolling circle amplification to detect mercuric ions. Nanomaterials 8, 81 (2018). https://doi.org/10.3390/nano8020081
Lim, J.W., Kim, T.Y., Lim, M.C., Choi, S.W., Woo, M.A.: Portable pumpless 3D-printed chip for on-site colorimetric screening of Hg2+ in lake water. BioChip J. 14, 169–178 (2020). https://doi.org/10.1007/s13206-019-4205-4
Kim, D., Lee, J., Park, S., Park, J., Seo, M.J., Rhee, W.J., Kim, E.: Target-responsive template structure switching-mediated exponential rolling circle amplification for the direct and sensitive detection of microRNA. BioChip J. (2022). https://doi.org/10.1007/s13206-022-00071-2
Lee, H.N., Lee, J., Kang, Y.K., Lee, J.H., Yang, S., Chung, H.J.: A lateral flow assay for nucleic acid detection based on rolling circle amplification using capture ligand-modified oligonucleotides. BioChip J. (2022). https://doi.org/10.1007/s13206-022-00080-1
Devadhasan, J.P., Kim, S.: CMOS image sensor based HIV diagnosis: a smart system for point-of-care approach. BioChip J. 7, 258–266 (2013). https://doi.org/10.1007/s13206-013-7309-2
Lim, J.W., Kim, T.Y., Choi, S.W., Woo, M.A.: 3D-printed rolling circle amplification chip for on-site colorimetric detection of inorganic mercury in drinking water. Food Chem. 300, 125177 (2019). https://doi.org/10.1016/j.foodchem.2019.125177
Han, Y.D., Chun, H.J., Yoon, H.C.: Low-cost point-of-care biosensors using common electronic components as transducers. BioChip J. 14, 32–47 (2020). https://doi.org/10.1007/s13206-020-4104-8
Kim, H.J., Kim, B.H., Seo, Y.H.: Immunoreaction-based microfluidic diagnostic device for the detection of prostate-specific antigen. BioChip J. 12, 154–162 (2018). https://doi.org/10.1007/s13206-017-2208-6
Jain, S., Nehra, M., Kumar, R., Dilbaghi, N., Hu, T., Kumar, S., Kaushik, A., Li, C.Z.: Internet of medical things (IoMT)-integrated biosensors for point-of-care testing of infectious diseases. Biosens. Bioelectron. 179, 113074 (2021). https://doi.org/10.1016/j.bios.2021.113074
Lim, J.M., Supianto, M., Kim, T.Y., Kim, B.S., Park, J.W., Jang, H.H., Lee, H.J.: Fluorescent lateral flow assay with carbon nanodot conjugates for carcinoembryonic antigen. BioChip J. (2023). https://doi.org/10.1007/s13206-022-00093-w
Lim, W.Y., Goh, C.H., Thevarajah, T.M., Goh, B.T., Khor, S.M.: Using SERS-based microfluidic paper-based device (μPAD) for calibration-free quantitative measurement of AMI cardiac biomarkers. Biosens. Bioelectron. 147, 111792 (2020). https://doi.org/10.1016/j.bios.2019.111792
Cho, H.H., Heo, J.H., Jung, D.H., Kim, S.H., Suh, S.J., Han, K.H., Lee, J.H.: Portable Au nanoparticle-based colorimetric sensor strip for rapid on-site detection of Cd2+ ions in potable water. BioChip J. 15, 276–286 (2021). https://doi.org/10.1007/s13206-021-00029-w
Nery, E.W., Kubota, L.T.: Sensing approaches on paper-based devices: a review. Anal. Bioanal. Chem. 405, 7573–7595 (2013). https://doi.org/10.1007/s00216-013-6911-4
Kuddannaya, S., Bao, J., Zhang, Y.: Enhanced in vitro biocompatibility of chemically modified poly (dimethylsiloxane) surfaces for stable adhesion and long-term investigation of brain cerebral cortex cells. ACS Appl. Mater. Interfaces 7, 25529–25538 (2015). https://doi.org/10.1021/acsami.5b09032
Miranda, I., Souza, A., Sousa, P., Ribeiro, J., Castanheira, E.M., Lima, R., Minas, G.: Properties and applications of PDMS for biomedical engineering: a review. J. Funct. Biomater. 13, 2 (2021). https://doi.org/10.3390/jfb13010002
Huyen, L.T.N., Hong, S.J., Trung, T.Q., Meeseepong, M., Kim, A.R., Lee, N.E.: Flexible capillary microfluidic devices based on surface-energy modified polydimethylsiloxane and polymethylmethacrylate with room-temperature chemical bonding. BioChip J. (2023). https://doi.org/10.1007/s13206-023-00096-1
Escobar, A., Chiu, P., Qu, J., Zhang, Y., Xu, C.Q.: Integrated microfluidic-based platforms for on-site detection and quantification of infectious pathogens: Towards on-site medical translation of SARS-CoV-2 diagnostic platforms. Micromachines 12, 1079 (2021). https://doi.org/10.3390/mi12091079
Trinh, K.T.L., Lee, N.Y.: Fabrication of wearable PDMS device for rapid detection of nucleic acids via recombinase polymerase amplification operated by human body heat. Biosensors 12, 72 (2022). https://doi.org/10.3390/bios12020072
Zhang, Y., Xu, J., Zhou, S., Zhu, L., Lv, X., Zhang, J., Zhang, L., Zhu, P., Yu, J.: DNAzyme-triggered visual and ratiometric electrochemiluminescence dual-readout assay for Pb (II) based on an assembled paper device. Anal. Chem. 92, 3874–3881 (2020). https://doi.org/10.1021/acs.analchem.9b05343
Fan, K., Zeng, J., Yang, C., Wang, G., Lian, K., Zhou, X., Deng, Y., Liu, G.: Digital quantification method for sensitive point-of-care detection of salivary uric acid using smartphone-assisted μPADs. ACS Sens. 7, 2049–2057 (2022). https://doi.org/10.1021/acssensors.2c00854
Acknowledgements
This work was carried out with the support of the Main Research Program (No. E0210701-03) of the Korea Food Research Institute, funded by the Ministry of Science and ICT, and the Cooperative Research Program for Agriculture Science and Technology Development (No. PJ016093), funded by Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
JHJ and M-AW conceived and designed the experiments; T-YK and SK performed the experiments; JHJ and M-AW analyzed and discussed the results; and T-YK and SK wrote the paper. All authors discussed the results and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Information:
Supplementary materials include the released test results of the RCA reaction solution from the PDMS microchip, sequence information of oligonucleotides used in this study, information on the paper chip, and urea-PAGE analysis results. The supplementary video shows the release of the reaction solution from the PDMS microchip to the paper chip using hand pressure (MP4, 119 MB). (DOCX 1580 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, TY., Kim, S., Jung, J.H. et al. Paper-Based Radial Flow Assay Integrated to Portable Isothermal Amplification Chip Platform for Colorimetric Detection of Target DNA. BioChip J 17, 263–273 (2023). https://doi.org/10.1007/s13206-023-00101-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-023-00101-7