Abstract
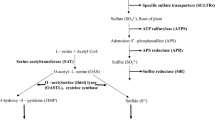
The biosynthesis of cysteine is crucial and critically regulated by two enzymes. i.e., serine acetyl transferase (SAT) and O-acetyl serine (thiol) lyase (OAS-TL). A descriptive account on the activity and regulatory mechanism of the enzyme is available in bacteria and plants. But no such studies yet performed in cyanobacteria, to understand the evolutionary aspect of cysteine biosynthesis and its regulation. Therefore, in our study a detailed bioinformatic analysis has been performed to understand all the possible features of cyanobacterial SATs and OAS-TLs. The analysis of SAT and OAS-TL sequences from cyanobacteria depicted that the large genome and morphological complexities favoured acquisition of these genes. Besides, conserved function of these enzymes was presumed by their sequence similarity. Further, the phylogenetic tree consisted of distinct clusters for unicellular, filamentous, and heterocytous strains. Nevertheless, the specificity pocket, SVKDR for OAS-TL having K as catalytic residue was also identified. Additionally, in silico protein modelling of SAT (SrpG) and OAS-TL (SrpH) of Synechococcus elongatus PCC 7942 was performed to gain insight into the structural attributes of the proteins. Finally, here we showed the possibility of hetero-oligomeric bi-enzyme cysteine synthase complex formation upon interaction of SAT and OAS-TL through protein–protein docking analysis thus provides a way to understand the regulation of cysteine biosynthesis in cyanobacteria.





Similar content being viewed by others
Data availability
Yes, the authors agreed for the availability of data to ensure data transparency norms.
References
Bairoch A, Bucher P, Hohann K (1996) The PROSITE database, its status in 1995. Nucleic Acids Res 24:189–196. https://doi.org/10.1093/nar/24.1.189
Berkowitz O, Wirtz M, Wolf A (2002) Use of bio molecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. J Biol Chem 277:30629–30634. https://doi.org/10.1074/jbc.M111632200
Bhattacharjee S, Mishra AK (2020) The tale of caspase homologues and their evolutionary outlook: deciphering programmed cell death in cyanobacteria. J Exp Bot 71:4639–4657. https://doi.org/10.1093/jxb/eraa213
Bochenek M, Etherington GJ, Koprivova A, Mugford ST, Bell TG, Malin G, Kopriva S (2013) Transcriptome analysis of the sulfate deficiency response in the marine microalga Emiliania huxleyi. New Phytol 199:650–662. https://doi.org/10.1111/nph.12303
Bogdanova N, Hell R (1997) Cysteine synthesis in plants: protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J 11:251–262. https://doi.org/10.1046/j.1365-313x.1997.11020251.x
Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56:187–220. https://doi.org/10.1146/annurev.arplant.56.032604.144246
Buchner P, Takahashi H, Hawkesford MJ (2004) Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J Exp Bot 55:1765–1773. https://doi.org/10.1093/jxb/erh206
Campanini B, Speroni F, Salsi E, Cook PF, Roderick SL, Huang B, Bettati S, Mozzarelli A (2005) Interaction of serine acetyltransferase with O-acetylserinesulfhydrylase active site: evidence from fluorescence spectroscopy. Protein Sci 14:2115–2124. https://doi.org/10.1110/ps.051492805
Chattopadhyay A, Meier M, Ivaninskii S, Burkhard P, Speroni F, Campanini B, Bettati S, Mozzarelli A, Rabeh WM, Li L, Cook PF (2007) Structure, mechanism, and conformational dynamics of O-acetylserine sulfhydrylase from Salmonella typhimurium: comparison of A and B isozymes. Biochemistry 46:8315–8330. https://doi.org/10.1021/bi602603c
Chinthalapudi K, Kumar M, Kumar S, Jain S, Ala N, Gourinath S (2008) Crystal structure of native O-acetyl-serine sulfhydrylase from Entamoeba histolytica and its complex with cysteine: Structural evidence for cysteine binding and lack of interactions with serine acetyl transferase. Proteins: Struct Funct Bioinform 72:1222–1232. https://doi.org/10.1002/prot.22013
Cook PF, Wedding RT (1976) A reaction mechanism from steady state kinetic studies for O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2. J Biol Chem 251:2023–2029. https://doi.org/10.1016/S0021-9258(17)33649-9
Davidian JC, Kopriva S (2010) Regulation of sulfate uptake and assimilation-the same or not the same? Mol Plant 3:314–325. https://doi.org/10.1093/mp/ssq001
Dicker IB, Seetharam S (1992) What is known about the structure and function of the Escherichia coli protein FirA? Mol Microbiol 6:817–823. https://doi.org/10.1111/j.1365-2958.1992.tb01532.x
Diessner W, Schmidt A (1980) Isoenzymes of cysteine synthase in the cyanobacterium Synechococcus 6301. Z Pjkmsenphysiol 102:57–68. https://doi.org/10.1016/S0044-328X(81)80217-6
Droux M, Ruffet ML, Douce R, Job D (1998a) Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants–structural and kinetic properties of the free and bound enzymes. Eur J Biochem 255:235–245. https://doi.org/10.1046/j.1432-1327.1998.2550235.x
Droux M, Ruffet ML, Douce R, Job D (1998b) Interactions between serine kinetic properties of the free and bound enzymes. Eur J Biochem 255:235–245. https://doi.org/10.1046/j.1432-1327.1998.2550235.x
Feldman-Salit A, Wirtz M, Hell R, Wade RC (2009) A mechanistic model of the cysteine synthase complex. J Mole Biol 386:37–59. https://doi.org/10.1016/j.jmb.2008.08.075
Feldman-Salit A, Wirtz M, Lenherr ED, Throm C, Hothorn M, Scheffzek K, Hell R, Wade RC (2012) Allosterically gated enzyme dynamics in the cysteine synthase complex regulate cysteine biosynthesis in Arabidopsis thaliana. Structure 20:292–302. https://doi.org/10.1016/j.str.2011.11.019
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Foyer CH, Noctor G (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155:2–18. https://doi.org/10.1104/pp.110.167569
Francois JA, Kumaran S, Jez JM (2006) Structural basis for interaction of O-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex. Plant Cell 18:3647–3655. https://doi.org/10.1105/tpc.106.047316
Gasteiger E, Hoogland C, Gattiker A (2005) Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. Humana press, pp 571–607
Gorman J, Shapiro L (2004) Structure of serine acetyltransferase from Haemophilus influenzae Rd. Acta Crystallogr Sect D Biol Crystallogr 60:1600–1605. https://doi.org/10.1107/S0907444904015240
Govardhana M, Kumudini BS (2020) In-silico analysis of cucumber (Cucumis sativus L.) Genome for WRKY transcription factors and cis-acting elements. Comp Biol Chem 85:107212. https://doi.org/10.1016/j.compbiolchem.2020.107212
Hopkins L, Parmar S, Blaszczyk A, Hesse H, Hoefgen R, Hawkesford MJ (2005) O-acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiol 138:433–440. https://doi.org/10.1104/pp.104.057521
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comp Appl Biosci 8:275–282. https://doi.org/10.1093/bioinformatics/8.3.275
Joshi P, Gupta A, Gupta V (2019) Insights into multifaceted activities of CysK for therapeutic interventions. 3 Biotech 9:44. https://doi.org/10.1007/s13205-019-1572-4
Jost R, Berkowitz O, Wirtz M, Hopkins L, Hawkesford MJ, Hell R (2000) Genomic and functional characterization of the oas gene family encoding O acetylserine (thiol)lyases, enzymes catalyzing the final step in cysteine biosynthesis in Arabidopsis thaliana. Gene 253:237–247. https://doi.org/10.1016/s0378-1119(00)00261-4
Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1522. https://doi.org/10.1038/nprot.2012.085
Khan MS, Haas FH, Allboje Samami A, Moghaddas Gholami A, Bauer A, Fellenberg K, Reichelt M, Hansch R, Mendel RR, Meyer AJ, Wirtz M, Hell R (2010) Sulfite reductase defines a newly discovered bottleneck for assimilatory sulfate reduction and is essential for growth and development in Arabidopsis thaliana. Plant Cell 22:1216–1231. https://doi.org/10.1105/tpc.110.074088
Kharwar S, Mishra AK (2020) Unraveling the complexities underlying sulfur deficiency and starvation in the cyanobacterium Anabaena sp. PCC 7120. Environ Exp Bot 172:103966. https://doi.org/10.1016/j.envexpbot.2019.103966
Kharwar S, Bhattacharjee S, Mishra AK (2021) Disentangling the impact of sulfur limitation on exopolysaccharide and functionality of Alr2882 by in silico approaches in Anabaena sp. PCC 7120. Appl Biochem Biotech. https://doi.org/10.1007/s12010-021-03501-3
Kopriva S, Koprivova A (2003) Sulphate assimilation: a pathway which likes to surprise. In: Sulphur in plants. Springer, Dordrecht, pp 87–112. https://doi.org/10.1007/978-94-017-0289-8_5
Kredich NM (1996) Biosynthesis of cysteine. In: Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umberger E (eds) Escherichia coli and Salmonella typhimurium. Cellular and molecular biology. ASM Press, Washington D.C., pp 514–527
Kredich NM, Tomkins GM (1966) The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem 241:4955–4965. https://doi.org/10.1016/S0021-9258(18)99657-2
Kredich NM, Becker MA, Tomkins GM (1969) Purification and characterization of cysteine synthetase, a bifunctional protein complex, from Salmonella typhimurium. J Biol Chem 244:2428–2439. https://doi.org/10.1016/S0021-9258(19)78241-6
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kumaran S, Jez JM (2007) Thermodynamics of the interaction between O-acetylserine sulfhydrylase and the C-terminus of serine acetyltransferase. Biochemistry 46:5586–5594. https://doi.org/10.1021/bi7001168
Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H (2001) Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 414:325–329. https://doi.org/10.1038/35104586
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol 8:477–486. https://doi.org/10.1007/BF00228148
Lehmann C, Begley TP, Ealick SE (2006) Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry 45:11–19. https://doi.org/10.1021/bi051502y
Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. https://doi.org/10.1093/nar/gkr201
Lovell SC, Davis IW, Arendall WB III, Lovell SC, Davis IW, Arendall WB III, De Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC (2002) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Struct Fun Genet 50:437–450. https://doi.org/10.1002/prot.10286
Marceau M, Lewis SD, Scaefer LA (1988a) The glycine-rich region of Escherichia coli D-serine dehydratase. J Bol Chem 263:16934–16941. https://doi.org/10.1016/S0021-9258(18)37481-7
Marceau M, McFali E, Lewis SD, Scaefer JA (1988b) D-serine dehydratase from Escherichia coli. J Biol Chem 263:16926–16933. https://doi.org/10.1016/j.bbapap.2011.10.017
Masada M, Fukushima K, Tamura G (1975) Cysteine synthase from rape leaves. J Biochem 77:1107–1115. https://doi.org/10.1093/oxfordjournals.jbchem.a130811
Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K (1999) Purification and characterization of serine acetyltransferase from Escherichia coli partially truncated at the C-terminal region. Biosci Biotech Biochem 63:168–179. https://doi.org/10.1271/bbb.63.168
Momany C, Ernst S, Hackert ML (1992) Ornithine decarboxylase from Lactobacillus embodies a PLP-binding scaffold and a GTP effector site. Am Crysiallogr Assoc Absir 20:44
Momany C, Ghosh R, Hackert ML (1995) Structural motifs for pyridoxal-5′-phosphate binding in decarboxylases: an analysis based on the crystal structure of the Lactobacillus 30a ornithine decarboxylase. Pro Sci 4:849–854. https://doi.org/10.1002/pro.5560040504
Mozzarelli A, Bettati S, Campanini B, Salsi E, Raboni S, Singh R, Spyrakis F, Kumar VP, Cook PF (2011) The multifaceted pyridoxal 5′-phosphate-dependent O-acetylserine sulfhydrylase. Biochim Biophys Acta 1814:1497–1510. https://doi.org/10.1016/j.bbapap.2011.04.011
Nicholson ML, Gaasenbeek M, Laudenbach DE (1995) Two enzymes together capable of cysteine biosynthesis are encoded on a cyanobacterial plasmid. Mol Gen Gene 247:623–632. https://doi.org/10.1007/BF00290354
Ning P, Liu C, Kong J, Lv J (2017) Genome-wide analysis of WRKY transcription factors in wheat (Triticum aestivum L.) and differential expression under water deficit condition. Peer J 5:e3232. https://doi.org/10.7717/peerj.3232
Noctor G, Mhamdi A, Chaouch S, Han YI, Neukermans J, Marquez-Garcia BE, Queval G, Foyer CH (2012) Glutathione in plants: an integrated overview. Plant Cell Environ 35:454–484. https://doi.org/10.1111/j.1365-3040.2011.02400.x
Olsen LR, Vetting MW, Roderick SL (2007) Structure of the E. coli bifunctional GlmU acetyltransferase active site with substrates and products. Protein Sci 16:1230–1235. https://doi.org/10.1110/ps.072779707
Rabeh WM, Cook PF (2004) Structure and mechanism of O-acetylserine sulfhydrylase. J Biol Chem 279:26803–26806. https://doi.org/10.1074/jbc.R400001200
Raj I, Kumar S, Gourinath S (2012) The narrow active-site cleft of O-acetylserine sulfhydrylase from Leishmania donovani allows complex formation with serine acetyltransferases with a range of C-terminal sequences. Acta Crystallogr Sect D Biol Crystallogr 68:909–919. https://doi.org/10.1107/S0907444912016459
Richau KH, Kaschani F, Verdoes M, Pansuriya TC, Niessen S, Stüber K, Colby T, Overkleeft HS, Bogyo M, Van der Hoorn RA (2012) Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol 158:1583–1599. https://doi.org/10.1104/pp.112.194001
Ruffet ML, Droux M, Douce R (1994) Purification and kinetic properties of serine acetyltransferase free of O-acetylserine(thiol)lyase spinach chloroplasts. Plant Physiol 104:597–604. https://doi.org/10.1104/pp.104.2.597
Saito K, Kurosawa M, Murakoshi I (1993) Determination of functional lysine residue of a plant cysteine synthase by site-directed mutagenesis, and the molecular evolutionary implications. FEBS 328:111–114. https://doi.org/10.1016/0014-5793(93)80976-2
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins: Struct Funct Bioinform 17:355–362. https://doi.org/10.1002/prot.340170404
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P (2016) The STRING database in 2017: quality controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw937
Takahashi H, Kopriva S, Giordano M, Saito K, Hell R (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Ann Rev Plant Biol 62:157–184. https://doi.org/10.1146/annurev-arplant-042110-103921
Vaara M (1992) Eight bacterial proteins, including UDP-N-acetylglucosamine acyltransferase (LpxA) and three other transferases of Escherichia coli, consist of a six-residue periodicity theme. FEMS Microbiol Lett 97:249–254. https://doi.org/10.1016/0378-1097(92)90344-n
Vijay-Kumar S, Bugg CE, Wilkinson KD, Cook WJ (1985) Three-dimensional structure of ubiquitin at 2.8 Å resolution. Proc Natl Acad Sci USA 82:3582–3585. https://doi.org/10.1073/pnas.82.11.3582
Vuorio R, Härkönen T, Tolvanen M, Vaara M (1994) The novel hexapeptide motif found in the acyltransferases LpxA and LpxD of lipid A biosynthesis is conserved in various bacteria. FEBS Lett 337:289–292. https://doi.org/10.1016/0014-5793(94)80211-4
Wheeler TJ, Clements J, Finn RD (2014) Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinform 15:7. https://doi.org/10.1186/1471-2105-15-7
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(suppl_2):W407–W410. https://doi.org/10.1093/nar/gkm290
Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS (2003) VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res 31:3316–3319. https://doi.org/10.1093/nar/gkg565
Wirtz M, Hell R (2006) Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. J Plant Physiol 163:273–286. https://doi.org/10.1016/j.jplph.2005.11.013
Wirtz M, Berkowitz O, Droux M, Hell R (2001) The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction. Eur J Biochem 268:686–693. https://doi.org/10.1046/j.1432-1327.2001.01920.x
Yi H, Galant A, Ravilious GE, Preuss ML, Jez JM (2010) Sensing sulphur conditions: simple to complex protein regulatory mechanisms in plant thiol metabolism. Mol Plant 3:269–279. https://doi.org/10.1093/mp/ssp112
Zhang C, Freddolino PL, Zhang Y (2017) COFACTOR: improved protein function prediction by combining structure, sequence, and protein-protein interaction information. Nucleic Acids Res 45:W291–W299. https://doi.org/10.1093/nar/gkx366
Acknowledgements
Surbhi Kharwar wants to thank University Grants Commission (UGC), New Delhi, India, for financial grant and also grateful to Dr. Vinay Kumar Singh, Information Officer, School of Biotechnology, Banaras Hindu University, Varanasi, India for providing molecular docking facility by Biovia Discovery Studio 2019. Samujjal Bhattacharjee is thankful to Council of Scientific and Industrial Research (CSIR), New Delhi, for awarding Junior Research Fellowship. The authors would also like to take this opportunity to thank Head, Department of Botany, Banaras Hindu University for providing the necessary facilities and encouragement to carry out this work.
Author information
Authors and Affiliations
Contributions
SK and AKM conceived the idea and designed the experiments. SK conducted the experiments and wrote the manuscript. SK, SB, and AKM critically review the manuscript. The manuscript was approved by all the authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
All authors participant agree to publish this work in your esteem Journal: 3 Biotech.
Consent to publish
Yes, all authors have given their consent.
Supplementary Information
Below is the link to the electronic supplementary material.
13205_2021_2899_MOESM3_ESM.pdf
Supplementary file3 Fig. S1. Hidden Markov Model (HMM) logos of cyanobacterial SAT (a) and OAS-TL (b) proteins. Logos were built with Skylign based on the alignment of protein sequences. The height of each amino acid code for each position reflects the weighted probability of occurrence at that position. The first amino acids Val, Leu, or Ile, followed often by Gly in the hexapeptide repeats with a consensus sequence of {V/L/I}-G-X-X-X-X, are marked with red boxes in the classical SAT proteins sequences. The blue box indicates the PLP-binding residues in the protein sequences. The SAT binding sites were indicated by pink represent OAS binding sites. Fig. S2. Structural verification by VADAR analysis of SAT (a) and OAS-TL (b) proteins showing functional accessible surface area, functional residues volume, stero/packing quality index, and 3D profile quality index. Fig. S3. Interacting sites of the SAT and OAS-TL proteins. (PDF 3189 kb)
Rights and permissions
About this article
Cite this article
Kharwar, S., Bhattacharjee, S. & Mishra, A.K. Bioinformatics analysis of enzymes involved in cysteine biosynthesis: first evidence for the formation of cysteine synthase complex in cyanobacteria. 3 Biotech 11, 354 (2021). https://doi.org/10.1007/s13205-021-02899-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02899-1