Abstract
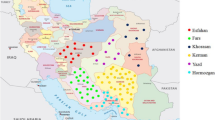
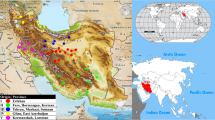
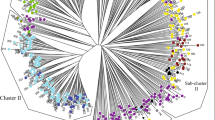
The importance of plant genetic resources has received increasing attention in crop improvement and plant variation in breeding procedures. Here, plant variation and the structures of plant populations were examined among 93 flax accessions using SCoT and URP molecular markers. Polymorphic-fragments by URP and SCoT primers were, on average, 9.3 and 10.9, respectively, and the mean of polymorphism information content for primers revealed a good efficiency of both marker techniques (0.34 and 0.35 for URP and SCoT, respectively). The neighbor-joining (NJ)-based clustering, via combined data, classified all investigated accessions into four main groups that were further confirmed by principal coordinate analysis (PCoA). Moreover, molecular diversity (88%) was mostly observed inside the populations. STRUCTURE-analysis confirmed the cluster analysis and all samples were separated into four subpopulations (∆K = 4). The results implied that the sub-populations consisted of diverse accessions, suggesting that this gene pool has the potential for flax breeding programs. Furthermore, our findings indicated that URP and SCoT markers are reliable techniques in assessing plant variation, especially in elucidating the structures of flax populations. However, gene-targeting markers like SCoT are preferable because of their roots in functional genome-based regions.







Similar content being viewed by others
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Abouseadaa HH, Atia MAM, Younis IY, Issa MY, Ashour HA, Saleh I, Osman GH, Arif IA, Engy M (2020) Gene-targeted molecular phylogeny, phytochemical profiling, and antioxidant activity of nine species belonging to family Cactaceae Saudi. J Biol Sci 27:1649–1658
Amirmoradi B, Talebi R, Karami E (2012) Comparison of genetic variation and differentiation among annual Cicer species using start codon targeted (SCoT) polymorphism, DAMDPCR, and ISSR markers. Plant Syst Evol 298:1679–1688
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Asadi A, Shooshtari L (2021) Assessment of somaclonal variation in micropropagation of Damask Rose (Rosa damascena Mill.) using molecular markers. MGJ 63:327–335
Bassett CM, Rodriguez-Leyva D, Pierce GN (2009) Experimental and clinical research findings on the cardiovascular benefits of consuming flaxseed. Appl Physiol Nutr Metab 34(5):965–974
Bhawna G, Abdin MZ, Arya L, Verma M (2017) Use of SCoT markers to assess the gene flow and population structure among two different populations of bottle gourd. Plant Gene 9:80–86
Chandrawati SN, Kumar R, Kumar S, Singh PK, Yadav VK, Ranade SA, Yadav HK (2017) Genetic diversity, population structure and association analysis in linseed (Linum usitatissimum L.). Physiol Mol Biol Plants 23:1–14
Cloutier S, Ragupathy R, Niu Z, Duguid S (2010) SSR- based linkage map of flax (Linum usitatissimum L.) and mapping of QTLs underlying fatty acid composition traits. Mol Breed 28:437–451
Collard BC, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol Report 27(1):86
Daneshmand H, Etminan AR, Qaderi A (2017) Diversity evaluation of Trigonella foenum-graecum populations using DNA markers and phytochemical characteristics. JMP 63:16
de Lucena CM, de Lucena RFP, Costa GM, Carvalho TKN, da Silva Costa GG, da Nóbrega Alves RR, Pereira DD, da Silva Ribeiro JE, Alves CAB, Quirino ZGM, Nunes EN (2013) Use and knowledge of Cactaceae in Northeastern Brazil. J Ethnobiol Ehnomed 9(1):62
Diederichsen A, Richards K (2003) Cultivated flax and the genus Linum L.: taxonomy and germplasm conservation. In: Muir AD, Westcott ND (eds) Flax: the genus Linum. CRC Press, London, pp 22–54
Ding Q, Li J, Wang F, Zhang Y, Li H, Zhang J, Li, H, Zhang J, Gao J (2015) Characterization and development of EST-SSRs by deep transcriptome sequencing in Chinese cabbage (Brassica rapa L ssp pekinensis). Int J Genom 2015: 473028
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Earl DA, vonHoldt BM (2012) Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
El Sayed AA, Ezzat SM, Mostafa SH, Zedan SZ, Abdel-Sattar E, El Tanbouly N (2018) Inter simple sequence repeat analysis of genetic diversity and relationship in four egyptian flaxseed genotypes. Pharm Res 10(2):166
Etminan A, Pour-Aboughadareh A, Mohammadi R, Ahmadi-Rad A, Noori A, Mahdavian Z, Moradi Z (2016) Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol Biotechnol Equip 30(6):1075–1081
Etminan A, Pour-Aboughadareh A, Mehrabi AA, Shooshtari L, Ahmadi-Rad A, Moradkhani H (2019) Molecular characterization of the wild relatives of wheat using CAAT-box derived polymorphism. Plant Biosyst 153(3):398–405
Everaert I, De Riek J, De Loose M, Van Waes J, Van Bockstaele E (2001) Most similar variety grouping for distinctness evaluation of flax and linseed (Linum usitatissimum L.) varieties by means of AFLP and morphological data. Plant Var Seeds 14(2): 69–87
Feng S, Yang T, Liu Z, Chen LV, Hou N, Wang Y, Wei A (2015) Genetic diversity and relationships of wild and cultivated Zanthoxylum germplasms based on sequence-related amplified polymorphism (SRAP) markers. Genet Resour Crop Evol 62:1193–1204
Feng SG, He RF, Jiang MY, Lu JJ, Shen XX, Lu JJ, Liu JJ, Wang ZA, Wang HZ (2016) Genetic diversity and relationships of medicinal Chrysanthemum morifolium revealed by start codon targeted (SCoT) markers. Sci Hortic 201:118–123
Feng S, Zhu Y, Yu C, Jiao K, Jiang M, Lu J, Shen C, Ying Q, Wang H (2018) Development of species-specific SCAR markers, based on a SCoT analysis, to Authenticate Physalis (Solanaceae) species. Front Genet 9:192
Fu YB, Peterson G, Diederichsen A, Richards KW (2002) RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet Resour Crop Evol 49:253–259
Fu YB, Rowland GG, Duguid SD, Richards KW (2003) RAPD analysis of 54 north American flax cultivars. Crop Sci 43(4):1510–1515
Ghobadi G, Etminan A, Mehrabi AM, Shooshtari L (2021) Molecular diversity analysis in hexaploid wheat (Triticum aestivum L.) and two Aegilops species (Aegilops crassa and Aegilops cylindrica) using CBDP and SCoT markers. J Genet Eng Biotechnol 19(1):56
Ghobadi-Namin L, Etminan A, Ghanavati F, Azizinezhad R, Abdollahi P (2021) Screening and selection of one hundred flax (Linum usitatissimum) accessions for yield production. J Nat Fibers. 19:7296–7304
Gogoi B, Wann SB, Saikia SP (2020) Comparative assessment of ISSR, RAPD, and SCoT markers for genetic diversity in Clerodendrum species of North East India. Mol Biol 47:7365–7377
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int 2015:431487–431487
Goyal A, Sharma V, Upadhyay N, Gill S, Sihag M (2014) Flax and flaxseed oil: an ancient medicine & modern functional food. J Food Sci Technol 51(9):1633–1653
Guo DL, Zhang JY, Liu CH (2012) Genetic diversity in some grape varieties revealed by SCoT analyses. Mol Biol Rep 39:5307–5313
Heidari P, Etminan A, Azizinezhad R, Khosroshahli M (2017) Genomic variation studies in durum wheat (Triticum turgidum ssp. durum) using CBDP, SCoT and ISSR markers. Indian J Genet Plant. 77(3):379–386
Hoque A, Fiedler JD, Rahman M (2020) Genetic diversity analysis of a flax (Linum usitatissimum L.) global collection. BMC Genom 21:557
Jana TK, Singh NK, Koundal KR, Sharma TR (2005) Genetic differentiation of charcoal rot pathogen, Macrophomina phaseolina, into specific groups using URP-PCR. Can J Microbiol 51:159–164
Jump AS, Penuelas J (2005) Running to stand still: adaptation and the response of plants to rapid climate change. Ecol 8(9):1010–1020
Kandan A, Akhtar J, Singh B, Dixit D, Chand D, Roy A, Rajkumar S, Agarwal PC (2014) Molecular diversity of Bipolaris oryzae infecting Oryza sativa in India. Phytoparasitica 43(1):5–14
Kang HW, Park DS, Par YJ, You CH, Lee BM, Eun MY, Go SJ (2002) Fingerprinting of diverse genomes using PCR with universal rice primers generated from repetitive sequence of Korean weedy rice. Mol Cells 13:281–287
Khodaee L, Azizinezhad R, Etminan A, Khosroshahi M (2021) Assessment of genetic diversity among Iranian Aegilops triuncialis accessions using ISSR, SCoT, and CBDP markers. J Genet Eng Biotechnol 19(1):5
Kumar P, Akhtar J, Kandan A, Singh B, Kiran R, Nair K, Dubey S (2018) Efficacy of URP and ISSR markers to determine diversity of indigenous and exotic isolates of Curvularia lunata. Indian Phytopathol 71:235–242
Kumari S, Nirala PRB, Rani N, Prasad BD (2017) Selection criteria of linseed (Linum usitatissimum L.) genotypes for seed yield traits through correlation and path coefficient analysis. J Oilseeds Res. 34(3):171–174
Kumari A, Paul S, Sharma V (2018) Genetic diversity analysis using RAPD and ISSR markers revealed discrete genetic makeup in relation to fibre and oil content in Linum usitatissimum L. genotypes. Nucl 61(1):45–53
Kurt O, Bozkurt D (2006) Effect of temperature and photoperiod on seedling emergence of flax (Linum usitatissimum L.). J Agron 5:541–545
Luo C, He XH, Chen H, Ou SJ, Gao MP (2010) Analysis of diversity and relationships among mango cultivars using Start Codon Targeted (SCoT) markers. Biochem System Ecol 38(6):1176–1184
Mehrparvar M, Mohammadi Goltapeh E, Safaie N (2012) Evaluation of genetic diversity of Iranian Lecanicillium fungicola isolates using URP marker. J Crop Prot 1(3):229–238
Mhiret WN, Heslop-Harrison JS (2018) Biodiversity in Ethiopian linseed (Linum usitatissimum L.): molecular characterization of landraces and some wild species. Genet Resour Crop Evol 65(6):1603–1614
Mishra SK, Nag A, Naik A, Rath SC, Gupta K, Gupta AK, Panigrahi J (2019) Characterization of host response to bruchids (Callosobruchus chinensis and C. maculatus) in 39 genotypes belongs to 12 Cajanus spp. and assessment of molecular diversity inter se. J Stored Prod Res 81:76–90
Mohammadi SA, Prasanna BM (2003) Analysis of genetic diversity in crop plants-salient statistical tools and considerations. Crop Sci 43(4):1235–1248
Mostafavi A, Omidi M, Azizinezhad R, Etminan A, Naghdi BH (2021) Genetic diversity analysis in a mini core collection of Damask rose (Rosa damascena Mill.) germplasm from Iran using URP and SCoT markers. J Genet Eng Biotechnol 19:144
Mulpuri S, Muddanuru T, Francis G (2013) Start codon targeted (SCoT) polymorphism in toxic and non-toxic accessions of Jatropha curcas L. and development of a codominant SCAR marker. Plant Sci 207:117–127
Nowak JL, Okon S, Wieremczuk A (2020) Molecular diversity analysis of genotypes from four Aegilops species based on retrotransposon–microsatellite amplifed polymorphism (REMAP) markers. Cereal Res Commun 49(1):37–44
Pakseresht F, Talebi R, Karami E (2013) Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of landrace chickpea (Cicer arietinum L.) genotypes collected from north-west of Iran. Physiol Mol Biol Plants 19(4):563–574
Patial R, Paul S, Sharma D, Sood VK, Kumar N (2019) Morphological characterization and genetic diversity of linseed (Linum usitatissimum L.). J Oilseeds Res. 36(1):8–16
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Perrier X, Jacquemoud-Collet J (2006) DARwin software at available at: http://darwin.cirad.fr/
Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M (2017) Assessment of genetic diversity among Iranian Triticum germplasm using agro-morphological traits and start codon targeted (SCoT) markers. Cereal Res Commun 45(4):574–586
Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M (2018) Insight into the genetic variability analysis and relationships among some Aegilops and Triticum species, as genome progenitors of bread wheat, using SCoT markers. Plant Biosyst 152:694–703
Pour-Aboughadareh A, Poczai P, Etminan A, Jadidi O, Kianersi F, Shooshtari L (2022) An analysis of genetic variability and population structure in wheat germplasm using microsatellite and gene-based markers. Plants 11:1205
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Qaderi A, Omidi M, Pour-Aboughadareh A, Poczai P, Shaghani J, Mehrafarin A, Nohooji M, Etminan A (2019) Molecular diversity and phytochemical variability in the Iranian poppy (Papaver bracteatum Lindl.): a baseline for conservation and utilization in future breeding programmes. Ind Crops Prod 130:237–247
Que YX, Pan YB, Lu YH, Yang C, Yang Y, Huang N, Xu L (2014) Genetic analysis of diversity within a Chinese local sugarcane germplasm based on start codon targeted polymorphism. Bio Med Res Int 2014:1–10
Salahlou R, Safaie N, Shamsbakhsh M (2019) Using ISSR and URP-PCR markers in detecting genetic diversity among Macrophomina phaseolina isolates of sesame in Iran. J Crop Prot 8(3):293–309
Saroha A, Pal D, Kaur V, Kumar Bartwal A, Aravind J, Radhamani J, Rajkumar S, Kumar R, Sunil S, Gomashe SA, Wankhede DP (2022) Agro-morphological variability and genetic diversity in linseed (Linum usitatissimum L.) germplasm accessions with emphasis on flowering and maturity time. Genet Resour Crop Evol 69:315–333
Shaygan N, Etminan A, Majidi Hervan E, Azizinezhad R, Mohammadi R (2021) The study of genetic diversity in a minicore collection of durum wheat genotypes using agro-morphological traits and molecular markers. Cereal Res Commun 49(1):141–147
Soto-Cerda BJ, Diederichsen A, Ragupathy R, Cloutier S (2013) Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol 13(1):78
Talebi R, Nosrati S, Etminan A, Naji AM (2018) Genetic diversity and population structure analysis of landrace and improved safflower (Cartamus tinctorious L.) germplasm using arbitrary functional gene-based molecular markers. Biotechnol Biotechnol Equip 32(5):1183–1194
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thakur R, Paul S, Thakur G, Kumar A, Dogra R (2021) Assessment of genetic diversity among linseed (Linum usitatissimum L.) germplasm based on morphological traits. J Pharmacogn Phytochem 9(6):1785–1790
Uysal H, Fu YB, Kurt O, Peterson GW, Diederichsen A, Kusters P (2010) Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne Mill.) as revealed by ISSR markers. Genet Resour Crop Evol 57:1109–1119
Vinu V, Singh N, Vasudev S, Kumar Yadava D, Kumar S, Naresh S, Ramachandra Bhat S, Vinod PK (2013) Assessment of genetic diversity in Brassica juncea (Brassicaceae) genotypes using phenotypic differences and SSR markers. Rev Biol Trop 61(4):1919–1934
Xiong FQ, Zhong RC, Han ZQ, Jiang J, He LQ, Zhuang WJ, Tang R (2011) Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol Biol Rep 38:3487–3494
Yadav HK, Chandrawati D, Singh N, Kumar R, Kumar S, Ranade SA (2018) Agromorphological traits and microsatellite markers based genetic diversity in Indian genotypes of linseed (Linum usitatissimum L.). J Agric Sci Tech 19:707–718
Zhao X, Zou G, Zhao J, Hu L, Lan Y, He J (2020) Genetic relationships and diversity among populations of Paris polyphylla assessed using SCoT and SRAP markers. Physiol Mol Biol Plants 26:1281–1293
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
AE, LG and FG conceived the research idea and designed the experiments. LG and AE performed the experiments and analyzed the data. LG, RA and PA wrote the manuscript. AE and RA revised and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghobadi-Namin, L., Etminan, A., Ghanavati, F. et al. Analysis of genetic diversity and population structure in a mini core collection of flax (Linum usitatissimum L.) based on URP and SCoT markers. J. Crop Sci. Biotechnol. 27, 43–55 (2024). https://doi.org/10.1007/s12892-023-00210-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-023-00210-7