Abstract
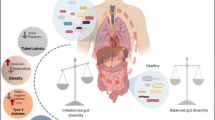
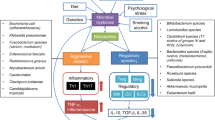
Inflammatory bowel disease (IBD) is an immune mediated chronic inflammatory disorder of gastrointestinal tract, which has underlying multifactorial pathogenic determinants such as environmental factors, susceptibility genes, gut microbial dysbiosis and a dysregulated immune response. Human gut is a frequent inhabitant of complex microbial ecosystem encompassing bacteria, viruses, parasites, fungi and other microorganisms that have an undisputable role in maintaining balanced homeostasis. All of these microbes interact with immune system and affect human gut physiology either directly or indirectly with interaction of each other. Intestinal fungi represent a smaller but crucial component of the human gut microbiome. Besides interaction with bacteriome and virome, it helps in balancing homoeostasis between pathophysiological and physiological processes, which is often dysregulated in patients with IBD. Understanding of gut mycobiome and its clinical implications are still in in its infancy as opposed to bacterial component of gut microbiome, which is more often focused. Modulation of gut mycobiome represents a novel and promising strategy in the management of patients with IBD. Emerging mycobiome-based therapies such as diet interventions, fecal microbiota transplantation (FMT), probiotics (both fungal and bacterial strains) and antifungals exhibit substantial effects in calibrating the gut mycobiome and restoring dysbalanced immune homeostasis by restoring the core gut mycobiome. In this review, we summarized compositional and functional diversity of the gut mycobiome in healthy individuals and patients with IBD, gut mycobiome dysbiosis in patients with IBD, host immune-fungal interactions and therapeutic role of modulation of intestinal fungi in patients with IBD.





Similar content being viewed by others
References
Silverberg MS, Satsangi J, Ahmad T, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19 Suppl A:5A-36A.
Ng SC, Bernstein CN, Vatn MH, et al. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut. 2013;62:630–49.
Benjamin JL, Hedin CR, Koutsoumpas A, et al. Smokers with active Crohn’s disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm Bowel Dis. 2012;18:1092–100.
Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3.
Wang M, Ahrné S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54:219–31.
Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8.
Iliev ID, Cadwell K. Effects of intestinal fungi and viruses on immune responses and inflammatory bowel diseases. Gastroenterology. 2021;160:1050–66.
Richard ML, Lamas B, Liguori G, Hoffmann TW, Sokol H. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:656–65.
Kapitan M, Niemiec MJ, Steimle A, Frick JS, Jacobsen ID. Fungi as part of the microbiota and interactions with intestinal bacteria. Curr Top Microbiol Immunol. 2019;422:265–301.
Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80.
Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65.
Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–16.
Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–91.
Schei K, Avershina E, Øien T, et al. Early gut mycobiota and mother-offspring transfer. Microbiome. 2017;5:107.
Liguori G, Lamas B, Richard ML, et al. Fungal dysbiosis in mucosa-associated microbiota of Crohns disease patients. J Crohn’s Colitis. 2016;10:296–305.
Nash AK, Auchtung TA, Wong MC, et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome. 2017;5:153.
Hoffmann C, Dollive S, Grunberg S, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8:e66019.
Wu L, Zeng T, Deligios M, et al. Age-related variation of bacterial and fungal communities in different body habitats across the young, elderly, and centenarians in Sardinia. Msphere. 2020;5:e00558-19.
Lai S, Yan Y, Pu Y, et al. Enterotypes of the human gut mycobiome. Microbiome. 2023;11:179.
Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–48.
Cao Y, Wang L, Ke S, et al. Analysis of intestinal mycobiota of patients with Clostridioides difficile infection among a prospective inpatient cohort. Microbiol Spectr. 2022;10:e0136222.
Zhang L, Zhan H, Xu W, Yan S, Ng SC. The role of gut mycobiome in health and diseases. Therap Adv Gastroenterol. 2021;14:17562848211047130.
Jeziorek M, Frej-Mądrzak M, Choroszy-Król I. The influence of diet on gastrointestinal Candida spp. colonization and the susceptibility of Candida spp. to antifungal drugs. Rocz Panstw Zakl Hig. 2019;70:195–200.
David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63.
Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–7.
Rosenbaum J, Usyk M, Chen Z, et al. Evaluation of oral cavity DNA extraction methods on bacterial and fungal microbiota. Sci Rep. 2019;9:1531.
Plato A, Hardison SE, Brown GD. Pattern recognition receptors in antifungal immunity. Semin Immunopathol. 2015;37:97–106.
Limon JJ, Tang J, Li D, et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe. 2019;25:377-88.e6.
Debelius J, Song SJ, Vazquez-Baeza Y, Xu ZZ, Gonzalez A, Knight R. Tiny microbes, enormous impacts: what matters in gut microbiome studies? Genome Biol. 2016;17:217.
Huseyin CE, Rubio RC, O’Sullivan O, Cotter PD, Scanlan PD. The fungal frontier: a comparative analysis of methods used in the study of the human gut mycobiome. Front Microbiol. 2017;8:1432.
Suhr MJ, Hallen-Adams HE. The human gut mycobiome: pitfalls and potentials—a mycologist’s perspective. Mycologia. 2015;107:1057–73.
Angebault C, Ghozlane A, Volant S, Botterel F, d’Enfert C, Bougnoux ME. Combined bacterial and fungal intestinal microbiota analyses: impact of storage conditions and DNA extraction protocols. PLoS One. 2018;13:e0201174.
Fiedorová K, Radvanský M, Němcová E, et al. The impact of DNA extraction methods on stool bacterial and fungal microbiota community recovery. Front Microbiol. 2019;10:821.
Sugiyama M, Xie XY, Atomi Y, Saito M. Differential diagnosis of small polypoid lesions of the gallbladder: the value of endoscopic ultrasonography. Ann Surg. 1999;229:498–504.
Tedersoo L, Anslan S, Bahram M, et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys. 2015;10:1–43.
Barfod KK, Poulsen SS, Hammer M, Larsen ST. Sub-chronic lung inflammation after airway exposures to Bacillus thuringiensis biopesticides in mice. BMC Microbiol. 2010;10:233.
Nilsson RH, Anslan S, Bahram M, Wurzbacher C, Baldrian P, Tedersoo L. Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat Rev Microbiol. 2019;17:95–109.
Tiew PY, Mac Aogain M, Ali NABM, et al. The mycobiome in health and disease: emerging concepts, methodologies and challenges. Mycopathologia. 2020;185:207–31.
Lindahl BD, Nilsson RH, Tedersoo L, et al. Fungal community analysis by high-throughput sequencing of amplified markers–a user’s guide. New Phytol. 2013;199:288–99.
Gao B, Chi L, Zhu Y, et al. An introduction to next generation sequencing bioinformatic analysis in gut microbiome studies. Biomolecules. 2021;11:530.
Aguiar-Pulido V, Huang W, Suarez-Ulloa V, Cickovski T, Mathee K, Narasimhan G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol Bioinform Online. 2016;12 Suppl 1:5–16.
Marcelino VR, Irinyi L, Eden JS, Meyer W, Holmes EC, Sorrell TC. Metatranscriptomics as a tool to identify fungal species and subspecies in mixed communities–a proof of concept under laboratory conditions. IMA Fungus. 2019;10:12.
Thielemann N, Herz M, Kurzai O, Martin R. Analyzing the human gut mycobiome - A short guide for beginners. Comput Struct Biotechnol J. 2022;20:608–14.
Bokulich NA, Subramanian S, Faith JJ, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–9.
Flynn JM, Brown EA, Chain FJ, MacIsaac HJ, Cristescu ME. Toward accurate molecular identification of species in complex environmental samples: testing the performance of sequence filtering and clustering methods. Ecol Evol. 2015;5:2252–66.
Westcott SL, Schloss PD. De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ. 2015;3:e1487.
Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 1994;44:846–9.
Kõljalg U, Nilsson RH, Abarenkov K, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7.
Nilsson RH, Hyde KD, Pawłowska J, et al. Improving ITS sequence data for identification of plant pathogenic fungi. Fungal Diversity. 2014;67:11–9.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7.
Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–41.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Wijayawardene NN, Bahram M, Sánchez-Castro I, et al. Current insight into culture-dependent and culture-independent methods in discovering Ascomycetous Taxa. J Fungi (Basel). 2021;7:703.
Soeta N, Terashima M, Gotoh M, et al. An improved rapid quantitative detection and identification method for a wide range of fungi. J Med Microbiol. 2009;58 Pt 8:1037–44.
Hoarau G, Mukherjee PK, Gower-Rousseau C, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio. 2016;7:e01250–16. https://doi.org/10.1128/mbio.01250-16.
Kombrink A, Tayyrov A, Essig A, et al. Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J. 2019;13:588–602.
Fernández de Ullivarri M, Arbulu S, Garcia-Gutierrez E, Cotter PD. Antifungal peptides as therapeutic agents. Front Cell Infect Microbiol. 2020;10:105.
Lambooij JM, Hoogenkamp MA, Brandt BW, Janus MM, Krom BP. Fungal mitochondrial oxygen consumption induces the growth of strict anaerobic bacteria. Fungal Genet Biol. 2017;109:1–6.
Noverr MC, Huffnagle GB. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect Immun. 2004;72:6206–10.
García C, Tebbji F, Daigneault M, et al. The human gut microbial metabolome modulates fungal growth via the TOR signaling pathway. mSphere. 2017;2:e00555–17. https://doi.org/10.1128/msphere.00555-17.
Zhang F, Aschenbrenner D, Yoo JY, Zuo T. The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe. 2022;3:e969–83.
Hsu C, Ghannoum M, Cominelli F, et al. Mycobiome and inflammatory bowel disease: role in disease pathogenesis, current approaches and novel nutritional-based therapies. Inflamm Bowel Dis. 2023;29:470–9.
Ott SJ, Kühbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–41.
Li XV, Leonardi I, Putzel GG, et al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature. 2022;603:672–8.
de Groot PW, Bader O, de Boer AD, Weig M, Chauhan N. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell. 2013;12:470–81.
Doron I, Mesko M, Li XV, et al. Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in Crohn’s disease. Nat Microbiol. 2021;6:1493–504.
Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–28.
Moyes DL, Wilson D, Richardson JP, et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 2016;532:64–8.
Leonardi I, Li X, Semon A, et al. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science. 2018;359:232–6.
Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8:352–8.
Sougioultzis S, Simeonidis S, Bhaskar KR, et al. Saccharomyces boulardii produces a soluble anti-inflammatory factor that inhibits NF-κB-mediated IL-8 gene expression. Biochem Biophys Res Commun. 2006;343:69–76.
Dahan S, Dalmasso G, Imbert V, Peyron JF, Rampal P, Czerucka D. Saccharomyces boulardii interferes with enterohemorrhagic Escherichia coli-induced signaling pathways in T84 cells. Infect Immun. 2003;71:766–73.
Dalmasso G, Cottrez F, Imbert V, et al. Saccharomyces boulardii inhibits inflammatory bowel disease by trapping T cells in mesenteric lymph nodes. Gastroenterology. 2006;131:1812–25.
Jawhara S, Poulain D. Saccharomyces boulardii decreases inflammation and intestinal colonization by Candida albicans in a mouse model of chemically-induced colitis. Med Mycol. 2007;45:691–700.
Underhill DM, Braun J. Fungal microbiome in inflammatory bowel disease: a critical assessment. J Clin Invest. 2022;132:e155786.
Chehoud C, Albenberg LG, Judge C, et al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:1948–56.
Li Q, Wang C, Tang C, He Q, Li N, Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J Clin Gastroenterol. 2014;48:513–23.
Jain U, Ver Heul AM, Xiong S, et al. Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science. 2021;371:1154–9.
Jun X, Ning C, Yang S, et al. Alteration of fungal microbiota after 5-ASA treatment in UC patients. Inflamm Bowel Dis. 2020;26:380–90.
Iliev ID. Mycobiota–host immune interactions in IBD: coming out of the shadows. Nat Rev Gastroenterol Hepatol. 2022;19:91–2.
Hsia K, Zhao N, Chung M, et al. Alterations in the fungal microbiome in ulcerative colitis. Inflamm Bowel Dis. 2023;29:1613–21.
Jangi S, Hsia K, Zhao N, et al. Dynamics of the gut mycobiome in patients with ulcerative colitis. Clin Gastroenterol Hepatol. 2023:S1542-3565(23)00762-0.
Krawczyk A, Salamon D, Kowalska-Duplaga K, et al. Changes in the gut mycobiome in pediatric patients in relation to the clinical activity of Crohn’s disease. World J Gastroenterol. 2023;29:2172–87.
Gross O, Poeck H, Bscheider M, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–6.
Hise AG, Tomalka J, Ganesan S, et al. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe. 2009;5:487–97.
Hager CL, Isham N, Schrom KP, et al. Effects of a novel probiotic combination on pathogenic bacterial-fungal polymicrobial biofilms. mBio. 2019;10:e00338–19.
Di Martino L, De Salvo C, Buela KA, et al. Candida tropicalis infection modulates the gut microbiome and confers enhanced susceptibility to colitis in mice. Cell Mol Gastroenterol Hepatol. 2022;13:901–23.
Li W, Shu Y, Zhang J, et al. Long-term prednisone treatment causes fungal microbiota dysbiosis and alters the ecological interaction between gut mycobiome and bacteriome in rats. Front Microbiol. 2023;14:1112767.
Yan P-G, Li J-N. Advances in the understanding of the intestinal micro-environment and inflammatory bowel disease. Chin Med J. 2020;133:834–41.
Wang S, Zhang Y-R, Yu Y-B. The important role of fungi in inflammatory bowel diseases. Scand J Gastroenterol. 2021;56:1312–22.
Glocker EO, Hennigs A, Nabavi M, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–35.
Schirbel A, Shouval DS, Hebecker B, Hube B, Sturm A, Werner L. Intestinal epithelial cells and T cells differentially recognize and respond to Candida albicans yeast and hypha. Eur J Immunol. 2018;48:1826–37.
Chiaro TR, Soto R, Zac Stephens W, et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017;9:eaaf9044.
Caër C, Wick MJ. Human intestinal mononuclear phagocytes in health and inflammatory bowel disease. Front Immunol. 2020;11:410.
Chikina AS, Nadalin F, Maurin M, et al. Macrophages maintain epithelium integrity by limiting fungal product absorption. Cell. 2020;183:411–28.e16.
Di Paola M, Rizzetto L, Stefanini I, et al. Comparative immunophenotyping of Saccharomyces cerevisiae and Candida spp. strains from Crohn’s disease patients and their interactions with the gut microbiome. J Transl Autoimmun. 2020;3:100036.
Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6:67–78.
Zelante T, De Luca A, Bonifazi P, et al. IL-23 and the Th17 pathway promote inflammation and impair antifungal immune resistance. Eur J Immunol. 2007;37:2695–706.
Duarte-Silva M, Afonso PC, de Souza PR, et al. Reappraisal of antibodies against Saccharomyces cerevisiae (ASCA) as persistent biomarkers in quiescent Crohn’s disease. Autoimmunity. 2019;52:37–47.
Pérez T, Balcázar JL, Ruiz-Zarzuela I, et al. Host–microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010;3:355–60.
Seow CH, Stempak JM, Xu W, et al. Novel anti-glycan antibodies related to inflammatory bowel disease diagnosis and phenotype. Am J Gastroenterol. 2009;104:1426–34.
Ost KS, O’Meara TR, Stephens WZ, et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature. 2021;596:114–8.
Hallen-Adams HE, Kachman SD, Kim J, Legge RM, Martínez I. Fungi inhabiting the healthy human gastrointestinal tract: a diverse and dynamic community. Fungal Ecology. 2015;15:9–17.
Suhr MJ, Banjara N, Hallen-Adams HE. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol. 2016;62:209–15.
Gunsalus KT, Tornberg-Belanger SN, Matthan NR, Lichtenstein AH, Kumamoto CA. Manipulation of host diet to reduce gastrointestinal colonization by the opportunistic pathogen Candida albicans. mSphere. 2015;1:e00020–15. https://doi.org/10.1128/mSphere.00020-15
Kostovcikova K, Coufal S, Galanova N, et al. Diet rich in animal protein promotes pro-inflammatory macrophage response and exacerbates colitis in mice. Front Immunol. 2019;10:919.
Lam S, Zuo T, Ho M, Chan FKL, Chan PKS, Ng SC. Fungal alterations in inflammatory bowel diseases. Aliment Pharmacol Ther. 2019;50:1159–71.
Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18:489–500.
Sun Y, Zuo T, Cheung CP, et al. Population-level configurations of gut mycobiome across 6 ethnicities in urban and rural China. Gastroenterology. 2021;160:272-86. e11.
Tannock GW, Liu Y. Guided dietary fibre intake as a means of directing short-chain fatty acid production by the gut microbiota. J R Soc N Z. 2020;50:434–55.
Guinan J, Wang S, Hazbun TR, Yadav H, Thangamani S. Antibiotic-induced decreases in the levels of microbial-derived short-chain fatty acids correlate with increased gastrointestinal colonization of Candida albicans. Sci Rep. 2019;9:8872.
Ghannoum M, Smith C, Adamson E, Isham N, Salem I, Retuerto M. Effect of mycobiome diet on gut fungal and bacterial communities of healthy adults. J Prob Health. 2019;7:215.
Klassert TE, Hanisch A, Bräuer J, et al. Modulatory role of vitamin A on the C andida albicans-induced immune response in human monocytes. Med Microbiol Immunol. 2014;203:415–24.
Bouzid D, Merzouki S, Bachiri M, Ailane SE, Zerroug MM. Vitamin D3 a new drug against Candida albicans. J Mycol Med. 2017;27:79–82.
Xie J, Zhu L, Zhu T, et al. Zinc supplementation reduces Candida infections in pediatric intensive care unit: a randomized placebo-controlled clinical trial. J Clin Biochem Nutr. 2019;64:170–3.
Baunwall SMD, Lee MM, Eriksen MK, et al. Faecal microbiota transplantation for recurrent Clostridioides difficile infection: an updated systematic review and meta-analysis. EClinicalMedicine. 2020;29–30:100642.
Mullish BH, Quraishi MN, Segal JP, et al. The use of faecal microbiota transplant as treatment for recurrent or refractory Clostridium difficile infection and other potential indications: joint British Society of Gastroenterology (BSG) and Healthcare Infection Society (HIS) guidelines. Gut. 2018;67:1920–41.
Zuo T, Wong SH, Cheung CP, et al. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat Commun. 2018;9:3663.
Moayyedi P, Surette MG, Kim PT, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149:102-9. e6.
Paramsothy S, Kamm MA, Kaakoush NO, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–28.
Costello SP, Hughes PA, Waters O, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321:156–64.
Haifer C, Paramsothy S, Kaakoush NO, et al. Lyophilised oral faecal microbiota transplantation for ulcerative colitis (LOTUS): a randomised, double-blind, placebo-controlled trial. Lancet Gastroenterol Hepatol. 2022;7:141–51.
Tan XY, Xie YJ, Liu XL, Li XY, Jia B. A systematic review and meta-analysis of randomized controlled trials of fecal microbiota transplantation for the treatment of inflammatory bowel disease. Evid Based Complement Alternat Med. 2022;2022:8266793.
Feng J, Chen Y, Liu Y, et al. Efficacy and safety of fecal microbiota transplantation in the treatment of ulcerative colitis: a systematic review and meta-analysis. Sci Rep. 2023;13:14494.
Weingarden AR, Vaughn BP. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes. 2017;8:238–52.
Lam S, Bai X, Shkoporov AN, et al. Roles of the gut virome and mycobiome in faecal microbiota transplantation. Lancet Gastroenterol Hepatol. 2022;7:472–84.
Leonardi I, Paramsothy S, Doron I, et al. Fungal trans-kingdom dynamics linked to responsiveness to fecal microbiota transplantation (FMT) therapy in ulcerative colitis. Cell Host Microbe. 2020;27:823-9.e3.
Standaert-Vitse A, Sendid B, Joossens M, et al. Candida albicanscolonization and ASCA in familial Crohn’s disease. Am J Gastroenterol. 2009;104:1745–53.
Mukhopadhya I, Hansen R, Meharg C, et al. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect. 2015;17:304–10.
Qiu X, Ma J, Jiao C, et al. Alterations in the mucosa-associated fungal microbiota in patients with ulcerative colitis. Oncotarget. 2017;8:107577–88.
El Mouzan MI, Korolev KS, Al Mofarreh MA, et al. Fungal dysbiosis predicts the diagnosis of pediatric Crohn’s disease. World J Gastroenterol. 2018;24:4510–6.
Imai T, Inoue R, Kawada Y, et al. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J Gastroenterol. 2019;54:149–59.
Qiu X, Zhao X, Cui X, et al. Characterization of fungal and bacterial dysbiosis in young adult Chinese patients with Crohn’s disease. Therap Adv Gastroenterol. 2020;13:1756284820971202.
Nelson A, Stewart CJ, Kennedy NA, et al. The Impact of NOD2 genetic variants on the gut mycobiota in Crohn’s disease patients in remission and in individuals without gastrointestinal inflammation. J Crohns Colitis. 2021;15:800–12.
Zeng L, Feng Z, Zhuo M, et al. Fecal fungal microbiota alterations associated with clinical phenotypes in Crohn’s disease in southwest China. PeerJ. 2022;10:e14260.
Tursi A, Brandimarte G, Papa A, et al. Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL# 3 as adjunctive to a standard pharmaceutical treatment: a double-blind, randomized, placebo-controlled study. Am J Gastroenterol. 2010;105:2218–27.
Sivananthan K, Petersen AM. Review of Saccharomyces boulardii as a treatment option in IBD. Immunopharmacol Immunotoxicol. 2018;40:465–75.
Canonici A, Pellegrino E, Siret C, et al. Saccharomyces boulardii improves intestinal epithelial cell restitution by inhibiting αvβ5 integrin activation state. PLoS One. 2012;7:e45047.
Sen S, Mansell TJ. Yeasts as probiotics: mechanisms, outcomes, and future potential. Fungal Genet Biol. 2020;137:103333.
Thomas S, Metzke D, Schmitz J, Dörffel Y, Baumgart DC. Anti-inflammatory effects of Saccharomyces boulardii mediated by myeloid dendritic cells from patients with Crohn’s disease and ulcerative colitis. Am J Physiol Gastrointest Liver Physiol. 2011;301:G1083-92.
Guslandi M, Giollo P, Testoni PA. A pilot trial of Saccharomyces boulardii in ulcerative colitis. Eur J Gastroenterol Hepatol. 2003;15:697–8.
Guslandi M, Mezzi G, Sorghi M, Testoni PA. Saccharomyces boulardii in maintenance treatment of Crohn’s disease. Dig Dis Sci. 2000;45:1462–4.
De Lourdes De Abreu Ferrari M, Sales Da Cunha A. Influence of Saccharomyces boulardii on the intestinal permeability of patients with Crohn’s disease in remission. Scand J Gastroenterol. 2008;43:842–8.
Bourreille A, Cadiot G, Le Dreau G, et al. Saccharomyces boulardii does not prevent relapse of Crohn’s disease. Clin Gastroenterol Hepatol. 2013;11:982–7.
Ghannoum MA, McCormick TS, Retuerto M, et al. Evaluation of microbiome alterations following consumption of BIOHM, a novel probiotic. Curr Issues Mol Biol. 2021;43:2135–46.
Di Martino L, Osme A, Ghannoum M, Cominelli F. A novel probiotic combination ameliorates Crohn’s disease–like ileitis by increasing short-chain fatty acid production and modulating essential adaptive immune pathways. Inflamm Bowel Dis. 2023;29:1105–17.
Huo X, Li D, Wu F, et al. Cultivated human intestinal fungus Candida metapsilosis M2006B attenuates colitis by secreting acyclic sesquiterpenoids as FXR agonists. Gut. 2022;71:2205–17.
Larsen IS, Jensen BAH, Bonazzi E, et al. Fungal lysozyme leverages the gut microbiota to curb DSS-induced colitis. Gut Microbes. 2021;13:1988836.
Scott BM, Gutiérrez-Vázquez C, Sanmarco LM, et al. Self-tunable engineered yeast probiotics for the treatment of inflammatory bowel disease. Nat Med. 2021;27:1212–22.
Bhaskaran N, Quigley C, Paw C, et al. Role of short chain fatty acids in controlling tregs and immunopathology during mucosal infection. Front Microbiol. 2018;9:1995.
Lührs H, Gerke T, Müller JG, et al. Butyrate inhibits NF-κB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol. 2002;37:458–66.
Jena A, Dutta U, Shah J, et al. Oral fluconazole therapy in patients with active ulcerative colitis who have detectable candida in the stool: a double-blind randomized placebo-controlled trial. J Clin Gastroenterol. 2022;56:705–11.
Sendid B, Salvetat N, Sarter H, et al. A pilot clinical study on post-operative recurrence provides biological clues for a role of Candida yeasts and fluconazole in Crohn’s disease. J Fungi (Basel). 2021;7:324.
Zwolińska-Wcisło M, Sliwowski Z, Drozdowicz D, et al. Candidiasis in the experimental model of ulcerative colitis. Folia Med Cracov. 2007;48:71–84.
Author information
Authors and Affiliations
Contributions
Amit Yadav: writing—original draft; writing—review and editing; Renu Yadav: writing—original draft; writing—review and editing; Vishal Sharma: writing—review and editing; Usha Dutta: writing—review and editing.
Corresponding author
Ethics declarations
Conflict of interest
AY, RY, VS and UD declare no competing interests.
Disclaimer
The authors are solely responsible for the data and the content of the paper. In no way, the Honorary Editor-in-Chief, Editorial Board Members, the Indian Society of Gastroenterology or printer/publishers are responsible for the results/findings and content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yadav, A., Yadav, R., Sharma, V. et al. A comprehensive guide to assess gut mycobiome and its role in pathogenesis and treatment of inflammatory bowel disease. Indian J Gastroenterol 43, 112–128 (2024). https://doi.org/10.1007/s12664-023-01510-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12664-023-01510-0