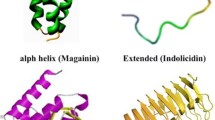
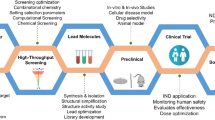
Abstract
The main issue with clinical infections is multidrug resistance to traditional antibiotics. As they are essential to innate immunity, shielding hosts from pathogenic microbes, traditional herbal remedies are an excellent supplier of antimicrobial peptides (AMPs), vital parts of defensive systems. Nevertheless, little is known about the bioactive peptide components of most ethnobotanical species. Our goal in this study was to find new, likely AMPs from Portulaca oleracea (P. oleracea) using in silico studies. The P. oleracea transcriptome was gained from Sequence Read Archive (SRA) and quality controlled, then adapters and other low-quality reads were trimmed. Afterward, de novo assembled and translated open reading frames (ORFs) were determined. Next, the ORFs were filtered based on AMP physiochemical criteria and deep learning methods. Finally, the five selected putative AMPs docked with E. coli and S. aureus membranes that showed penetration in bilayers. In this step, PO2 was chosen as a candidate AMP to analyze with molecular dynamics (MD) simulations. Our data demonstrated that PO2 is more stable in E. coli than in S. aureus. Moreover, these predicted AMPs can be good candidates for in vitro and in vivo analysis.








Similar content being viewed by others
Data Availability
The data generated or analyzed during this study are included in the manuscript and its supplementary information files.
References
Kesidis A, Depping P, Lodé A et al (2020) Expression of eukaryotic membrane proteins in eukaryotic and prokaryotic hosts. Methods 180:3–18. https://doi.org/10.1016/j.ymeth.2020.06.006
Moghaddam MM, Abolhassani F, Babavalian H, Mirnejad R, Azizi Barjini K, Amani J (2012) Comparison of in vitro antibacterial activities of two cationic peptides CM15 and CM11 against five pathogenic bacteria: Pseudomonas aeruginosa, Staphylococcus aureus, Vibrio cholerae, Acinetobacter baumannii, and Escherichia coli. Probiotics and Antimicrobial Proteins 4(2):133–139. https://doi.org/10.1007/s12602-012-9098-7
Moravej H, Moravej Z, Yazdanparast M et al (2018) Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb Drug Resist 24(6):747–767
Iranshahy M, Javadi B, Iranshahi M et al (2017) A review of traditional uses, phytochemistry and pharmacology of Portulaca oleracea L. J Ethnopharmacol 205:158–172. https://doi.org/10.1016/j.jep.2017.05.004
Zhou YX, Xin HL, Rahman K, Wang SJ, Peng C, Zhang H (2015) Portulaca oleracea L.: a review of phytochemistry and pharmacological effects. BioMed Research International 2015
Karimi G, Hosseinzadeh H, Ettehad N (2004) Evaluation of the gastric antiulcerogenic effects of Portulaca oleracea L. extracts in mice. Phytotherapy Research: An International Journal Devoted to Pharmacological and Toxicological Evaluation of Natural Product Derivatives 18 6:484–487. https://doi.org/10.1002/ptr.1463
Razi M. Al-Hawi fi'l-tibb (1968) Comprehensive book of medicine. Hyderabad: Osmania Oriental Publications Bureau 20:548–553.
Ibn SA, Al-Qānun fi’l-Ṭibb, (1987) Canon of medicine, in Arabic. IHMMR Printing Press, New Delhi
Ratledge C, Wilkinson S (1988) An overview of microbial lipids. Microbial Lipids 1:3–22
Ratledge C, Wilkinson S (1988) Fatty acids, related and derived lipids. Microbial Lipids 1:23–52
Bogdanov M, Dowhan W (1995) Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J Biol Chem 270(2):732–739
Domènech Ò, Torrent-Burgués J, Merino S, Sanz F, Montero MT, Hernández-Borrell J (2005) Surface thermodynamics study of monolayers formed with heteroacid phospholipids of biological interest. Colloids Surf, B 41(4):233–238. https://doi.org/10.1016/j.colsurfb.2004.12.012
Dickey A, Faller R (2008) Examining the contributions of lipid shape and headgroup charge on bilayer behavior. Biophys J 95(6):2636–2646. https://doi.org/10.1529/biophysj.107.128074
Epand RM, Epand RF (2009) Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochimica et Biophysica Acta (BBA)-Biomembranes 1788(1):289–294. https://doi.org/10.1016/j.bbamem.2008.08.023
Epand RM, Rotem S, Mor A, Berno B, Epand RF (2008) Bacterial membranes as predictors of antimicrobial potency. J Am Chem Soc 130(43):14346–14352
Leinonen R, Sugawara H, Shumway M (2010) The sequence read archive. Nucleic Acids Res 39(1):D19–D21. https://doi.org/10.1093/nar/gkq1019
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics 2010–2017.
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Grabherr MG, Haas BJ, Yassour M et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652
Singh U, Wurtele ES (2021) orfipy: a fast and flexible tool for extracting ORFs. Bioinformatics 37(18):3019–3020. https://doi.org/10.1093/bioinformatics/btab090
Bendtsen JD, Nielsen H, Von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340(4):783–795. https://doi.org/10.1016/j.jmb.2004.05.028
Pirtskhalava M, Amstrong AA, Grigolava M et al (2021) DBAASP v3: database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res 49(D1):D288–D297
Wang G, Li X, Wang Z (2016) APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44(D1):D1087–D1093
Vishnepolsky B, Grigolava M, Managadze G et al (2022) Comparative analysis of machine learning algorithms on the microbial strain-specific AMP prediction. Briefings in Bioinformatics 23(4):bbac233. https://doi.org/10.1093/bib/bbac233
Waghu FH, Idicula-Thomas S (2020) Collection of antimicrobial peptides database and its derivatives: applications and beyond. Protein Sci 29(1):36–42. https://doi.org/10.1002/pro.3714
Meher PK, Sahu TK, Saini V, Rao AR (2017) Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci Rep 7(1):42362
Jhong JH, Yao L, Pang Y et al (2022) dbAMP 2.0: updated resource for antimicrobial peptides with an enhanced scanning method for genomic and proteomic data. Nucleic Acids Research 50(D1):D460-D470. https://doi.org/10.1093/nar/gkab1080
Chaudhary K, Kumar R, Singh S et al (2016) A web server and mobile app for computing hemolytic potency of peptides. Sci Rep 6(1):22843. https://doi.org/10.1038/srep22843
Wei L, Ye X, Sakurai T, Mu Z, Wei L (2022) ToxIBTL: prediction of peptide toxicity based on information bottleneck and transfer learning. Bioinformatics 38(6):1514–1524. https://doi.org/10.1093/bioinformatics/btac006
Sharma A, Gupta P, Kumar R, Bhardwaj A (2016) dPABBs: a novel in silico approach for predicting and designing anti-biofilm peptides. Sci Rep 6(1):21839. https://doi.org/10.1038/srep21839
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519. https://doi.org/10.1002/pro.5560020916
Wu EL, Cheng X, Jo S et al (2014) CHARMM-GUI membrane builder toward realistic biological membrane simulations. J Comput Chem 35(27):1997–2004. https://doi.org/10.1002/jcc.23702
Zaeifi D, Najafi A, Mirnejad R (2023) Molecular dynamics simulation of antimicrobial peptide CM15 in Staphylococcus Aureus and Escherichia coli model bilayer lipid. Iran J Biotechnol 21(2):e3344
Allen WJ, Lemkul JA, Bevan DR (2009) GridMAT-MD: a grid-based membrane analysis tool for use with molecular dynamics. J Comput Chem 30(12):1952–1958. https://doi.org/10.1002/jcc.21172
Chen JE, Huang CC, Ferrin TE (2015) RRDistMaps: a UCSF Chimera tool for viewing and comparing protein distance maps. Bioinformatics 31(9):1484–1486. https://doi.org/10.1093/bioinformatics/btu841
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. Chemical Biology: Methods and Protocols 1263:243–250. https://doi.org/10.1007/978-1-4939-2269-7_19
Fantner GE, Barbero RJ, Gray DS, Belcher AM (2010) Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat Nanotechnol 5(4):280–285. https://doi.org/10.1038/nnano.2010.29
Abraham MJ, Murtola T, Schulz R et al (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25
Hub JS, De Groot BL, Van Der Spoel D (2010) g_wham-A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J Chem Theory Comput 6(12):3713–3720
Schrodinger L (2015) The PyMOL molecular graphics system Version 1:8
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Silva VNB, da Silva TLC, Ferreira TMM et al (2023) Multi-omics analysis of young Portulaca oleracea L. plants’ responses to high NaCl doses reveals insights into pathways and genes responsive to salinity stress in this halophyte species. Phenomics 3(1):1–21
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12(1):7–8. https://doi.org/10.1038/nmeth.3213
Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P (2016) PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 44(W1):W449–W454. https://doi.org/10.1093/nar/gkw329
Du Z, Su H, Wang W et al (2021) The trRosetta server for fast and accurate protein structure prediction. Nat Protoc 16(12):5634–5651. https://doi.org/10.1038/s41596-021-00628-9
Bungau S, Tit DM, Behl T, Aleya L, Zaha DC (2021) Aspects of excessive antibiotic consumption and environmental influences correlated with the occurrence of resistance to antimicrobial agents. Current Opinion in Environmental Science & Health 19:100224
Danis-Wlodarczyk K, Dąbrowska K, Abedon ST (2021) Phage therapy: the pharmacology of antibacterial viruses. Curr Issues Mol Biol 40(1):81–164. https://doi.org/10.21775/cimb.040.081
Mohammed I, Said DG, Dua HS (2017) Human antimicrobial peptides in ocular surface defense. Prog Retin Eye Res 61:1–22. https://doi.org/10.1016/j.preteyeres.2017.03.004
Lei J, Sun L, Huang S et al (2019) The antimicrobial peptides and their potential clinical applications. Am J Transl Res 11(7):3919
Lee TH, Hall NK, Aguilar MI (2016) Antimicrobial peptide structure and mechanism of action: a focus on the role of membrane structure. Curr Top Med Chem 16(1):25–39. https://doi.org/10.2174/1568026615666150703121700
Zhang QY, Yan ZB, Meng YM et al (2021) Antimicrobial peptides: mechanism of action, activity and clinical potential. Mil Med Res 8(1):1–25. https://doi.org/10.1186/s40779-021-00343-2
Moyer TB, Heil LR, Kirkpatrick CL et al (2019) PepSAVI-MS reveals a proline-rich antimicrobial peptide in Amaranthus tricolor. J Nat Prod 82(10):2744–2753. https://doi.org/10.1021/acs.jnatprod.9b00352
Shukurov RR, Voblikova DV, Nikonorova AK et al (2012) Transformation of tobacco and Arabidopsis plants with Stellaria media genes encoding novel hevein-like peptides increases their resistance to fungal pathogens. Transgenic Res 21:313–325
Khaliluev M, Kharchenko P, Dolgov S (2010) Genetic transformation of tomato (Solanum lycopersi cum L.) by protective chitin binding protein and antimicrobial peptide genes. Izv TSKhA 6:75–83
Wang W, Gu L, Dong L, Wang X (2007) Protective effect of Portulaca oleracea extracts on hypoxic nerve tissue and its mechanism. Asia Pac J Clin Nutr 16:227
Hozayen W, Bastawy M, Elshafeey H (2011) Effects of aqueous Purslane (Portulaca oleracea) extract and fish oil on gentamicin nephrotoxicity in albino rats. J Nat Sci 9(2):47–62
Karimi G, Aghasizadeh M, Razavi M, Taghiabadi E (2011) Protective effects of aqueous and ethanolic extracts of Nigella sativa L. and Portulaca oleracea L. on free radical induced hemolysis of RBCs. Daru: Journal of Faculty of Pharmacy, Tehran University of Medical Sciences 19(4):295.
Dan Z (2006) Study on antimicrobial effect of flavonoids from Portulace oleracea L. J Anhui Agric Sci 34(1):7
Eidi A, Mortazavi P, Moghadam JZ, Mardani PM (2015) Hepatoprotective effects of Portulaca oleracea extract against CCl4-induced damage in rats. Pharm Biol 53(7):1042–1051. https://doi.org/10.3109/13880209.2014.957783
Nayaka H, Londonkar RL, Umesh M (2014) Evaluation of potential antifertility activity of total flavonoids, isolated from Portulaca oleracea L on female albino rats. Int J PharmTech Res 6:783–793
Yakovlev IA, Lysøe E, Heldal I, Steen H, Hagen SB, Clarke JL (2020) Transcriptome profiling and in silico detection of the antimicrobial peptides of red king crab Paralithodes camtschaticus. Sci Rep 10(1):12679. https://doi.org/10.1038/s41598-020-69126-4
Seyedjavadi SS, Razzaghi-Abyaneh M, Nasiri MJ et al (2022) Isolation and chemical characterization of an alpha-helical peptide, dendrocin-ZM1, derived from zataria multiflora boiss with potent antibacterial activity. Probiotics and Antimicrobial Proteins 14(2):326–336. https://doi.org/10.1007/s12602-022-09907-7
Houyvet B, Zanuttini B, Corre E, Le Corguillé G, Henry J, Zatylny-Gaudin C (2018) Design of antimicrobial peptides from a cuttlefish database. Amino Acids 50(11):1573–1582. https://doi.org/10.1007/s00726-018-2633-4
Sanchez YBA, Agudelo M, Orduz S (2015) Peptide ID 1.0. Un programa para buscar potenciales pe´ptidos bioactivosen secuencias de proteinas. Dirección Nacional de Derechos de Autor. Ministerio del Interior. Registro 13–50–213 del 12 de Noviembre de 2015
Hincapié O, Giraldo P, Orduz S (2018) In silico design of polycationic antimicrobial peptides active against Pseudomonas aeruginosa and Staphylococcus aureus. Antonie Van Leeuwenhoek 111(10):1871–1882. https://doi.org/10.1007/s10482-018-1080-2
Barneh F, Nazarian A, Mousavi-nadushan R, Bagheri KP (2023) A novel in silico filtration method for discovery of encrypted antimicrobial peptides. Current Bioinformatics 1–11
Chakraborty A, Kobzev E, Chan J et al (2020) Molecular dynamics simulation of the interaction of two linear battacin analogs with model gram-positive and gram-negative bacterial cell membranes. ACS Omega 6(1):388–400. https://doi.org/10.1021/acsomega.0c04752
Talandashti R, Mehrnejad F, Rostamipour K et al (2021) Molecular insights into pore formation mechanism, membrane perturbation, and water permeation by the antimicrobial peptide pleurocidin: a combined all-atom and Coarse-Grained molecular dynamics simulation study. J Phys Chem B 125(26):7163–7176. https://doi.org/10.1021/acs.jpcb.1c01954
Acknowledgements
We thank the Engineering and Medical Physics Department for providing the computational server in bioinformatics analysis.
Author information
Authors and Affiliations
Contributions
BHA performed study design, in silico analysis, validation, and writing—original draft; SH, FA, FP, and AE performed in silico analysis and literature review; AB performed supervision, conceptualization, and manuscript review. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12602_2024_10261_MOESM1_ESM.jpg
Supplementary file1 (JPG 270 KB): Figure S1: Quality control of reads: (a and c) related to “Per base sequence content” analysis of forward and reverse reads before trimming, respectively; (b and d) related to “Per base sequence content” analysis of forward and reverse reads after trimming, respectively
12602_2024_10261_MOESM2_ESM.jpg
Supplementary file2 (JPG 25 KB): Figure S2: 3-D model of PO10 putative AMP: The residues at 10–14 positions (as shown in red color) make a coil structure and result in the rotation of helical structure
Supplementary file3 (MP4 6805 KB): Supplementary Video 1: PO2 movement along z-direction in E. coli (left side video) and S. aureus (right side video) systems: In both systems, the membrane and ions are depicted as spheres, while PO2 is represented as a new cartoon drawing method in red color.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hasannejad-Asl, B., Heydari, S., Azod, F. et al. Peptide-Membrane Docking and Molecular Dynamic Simulation of In Silico Detected Antimicrobial Peptides from Portulaca oleracea’s Transcriptome. Probiotics & Antimicro. Prot. (2024). https://doi.org/10.1007/s12602-024-10261-z
Accepted:
Published:
DOI: https://doi.org/10.1007/s12602-024-10261-z