Abstract
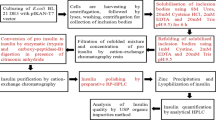
The plasminogen-free fibrin plate assay method was used to isolate Bacillus subtilis MX-6, a strain with high production of nattokinase from Chinese douchi. The presence of aprN, a gene-encoding nattokinase, was verified with PCR method. The predicted amino acid sequence was aligned with homologous sequences, and a phylogenetic tree was constructed. Nattokinase was sublimated with ammonium sulfate, using a DEAE-Sepharose Fast Flow column, a CM-Sepharose Fast Flow column and a Sephadex G-75 gel filtration column. SDS-PAGE analysis indicated that the molecular weight of the purified nattokinase from Bacillus subtilis MX-6 was about 28 kDa. Fermentation of Bacillus subtilis MX-6 nattokinase showed that nattokinase production was maximized after 72 h; the diameter of clear zone reached 21.60 mm on the plasminogen-free fibrin plate. Nattokinase production by Bacillus subtilis MX-6 increased significantly after supplementation with supernatant I, supernatant II and soy peptone but decreased substantially after the addition of amino acids. This result indicated that the nattokinase production by B. subtilis MX-6 might be induced by soybean polypeptides. The addition of MgSO4 and CaCl2 increased B. subtilis MX-6 nattokinase production.






Similar content being viewed by others
References
Cai D, Zhu C, Chen S (2017) Microbial production of nattokinase: current progress, challenge and prospect. World J Microb Biotechnol 33(5):84. https://doi.org/10.1007/s11274-017-2253-2
Wei X, Luo M, Xie Y, Yang L, Li H, Xu L, Liu H (2012) Strain screening, fermentation, separation, and encapsulation for production of nattokinase functional food. Appl Biochem Biotechnol 168(7):1753–1764. https://doi.org/10.1007/s12010-012-9894-2
Albuquerque W, Nascimento T, Brandão-Costa R, Fernandes T, Porto A (2017) Static magnetic field effects on proteases with fibrinolytic activity produced by Mucor subtilissimus. Bioelectromagnetics 38(2):109–120. https://doi.org/10.1002/bem.22016
Kapoor R, Panda BP (2013) Bioprospecting of soybean for production of nattokinase. Nutrafoods 12(3):89–95. https://doi.org/10.1007/s13749-013-0032-8
Mohanasrinivasan V, Mohanapriya A, Potdar S, Chatterji S, Konne S, Kumari S, Merlyn Keziah S, Subathra Devi C (2017) In vitro and in silico studies on fibrinolytic activity of nattokinase: a clot buster from Bacillus sp. Front Biol 12(3):219–225. https://doi.org/10.1007/s11515-017-1453-3
Toymentseva AA, Danilova IV, Tihonova AO, Sharipova MR, Balaban NP (2016) Purification of recombinant extracellular proteases from Bacillus pumilus for ß-amyloid peptide cleavage. Russ J Bioorg Chem 42(1):53–58. https://doi.org/10.1134/S1068162015060175
Chhikara A, Sharma S, Chandra J, Nangia A (2017) Thrombin activable fibrinolysis inhibitor in beta thalassemia. Indian J Pediatr 84(1):25–30. https://doi.org/10.1007/s12098-016-2208-x
Godier A, Parmar K, Manandhar K, Hunt BJ (2016) An in vitro study of the effects of t-PA and tranexamic acid on whole blood coagulation and fibrinolysis. J Clin Pathol 70:154–161
Zhang F, Zhang J, Linhardt RJ (2015) Interactions between nattokinase and heparin/GAGs. Glycoconj J 32(9):695–702. https://doi.org/10.1007/s10719-015-9620-8
Chen HS, Qi SH, Shen JG (2017) One-compound-multi-target: combination prospect of natural compounds with thrombolytic therapy in acute ischemic stroke. Curr Neuropharmacol 15(1):134–156
Singh R, Kumar M, Mittal A, Mehta PK (2016) Microbial enzymes: industrial progress in 21st century. 3 Biotech 6(2):174
Nguyen TT, Quyen TD, Le HT (2013) Cloning and enhancing production of a detergent- and organic-solvent-resistant nattokinase from Bacillus subtilis VTCC-DVN-12-01 by using an eight-protease-gene-deficient Bacillus subtilis WB800. Microb Cell Factories 12(1):79. https://doi.org/10.1186/1475-2859-12-79
Duan C, Feng Y, Zhou H, Xia X, Shang Y, Cui Y (2015) Optimization of fermentation condition of man-made bee-bread by response surface methodology. Adv Appl Microbiol 333:353–363
Vaithilingam M, Chandrasekaran SD, Gupta S, Paul D, Sahu P, Selvaraj JN, Babu V (2016) Extraction of Nattokinase enzyme from Bacillus cereus isolated from rust. Natl Acad Sci Lett 39(4):263–267. https://doi.org/10.1007/s40009-016-0476-7
Devi CS, Mohanasrinivasan V, Sharma P, Das D, Vaishnavi B, Naine SJ (2016) Production, purification and stability studies on nattokinase: a therapeutic protein extracted from mutant Pseudomonas aeruginosa CMSS isolated from bovine milk. Int J Pept Res Ther 22(2):263–269. https://doi.org/10.1007/s10989-015-9505-5
Dabbagh F, Negahdaripour M, Berenjian A, Behfar A, Mohammadi F, Zamani M, Ghasemi Y (2014) Nattokinase: production and application. Appl Microbiol Biotechnol 98(22):9199–9206. https://doi.org/10.1007/s00253-014-6135-3
Nagata C, Wada K, Tamura T, Konishi K, Goto Y, Koda S, Kawachi T, Tsuji M, Nakamura K (2016) Dietary soy and natto intake and cardiovascular disease mortality in Japanese adults: the Takayama study. Am J Clin Nutr 105:426–431
Wang C, Ji B, Li B, Ji H (2006) Enzymatic properties and identification of a fibrinolytic serine protease purified from Bacillus subtilis DC33. World J Microbiol Biotechnol 22(12):1365–1371. https://doi.org/10.1007/s11274-006-9184-7
Kotb E (2015) Purification and partial characterization of serine fibrinolytic enzyme from Bacillus megaterium KSK-07 isolated from kishk, a traditional Egyptian fermented food. Appl Biochem Microbiol 51(1):34–43. https://doi.org/10.1134/S000368381501007X
Anh DBQ, Mi NTT, Van Hung P (2015) Isolation and optimization of growth condition of Bacillus sp. from fermented shrimp paste for high fibrinolytic enzyme production. Arab J Sci Eng 40(1):23–28. https://doi.org/10.1007/s13369-014-1506-8
Thokchom S, Joshi SR (2014) Screening of fibrinolytic enzymes from lactic acid bacterial isolates associated with traditional fermented soybean foods. Food Sci Biotechnol 23(5):1601–1604. https://doi.org/10.1007/s10068-014-0217-y
Kotb E (2014) Purification and partial characterization of a chymotrypsin-like serine fibrinolytic enzyme from Bacillus amyloliquefaciens FCF-11 using corn husk as a novel substrate. World J Microbiol Biotechnol 30(7):2071–2080. https://doi.org/10.1007/s11274-014-1632-1
Lu F, Sun L, Lu Z, Bie X, Fang Y, Liu S (2007) Isolation and identification of an endophytic strain EJS-3 producing novel fibrinolytic enzymes. Curr Microbiol 54(6):435–439. https://doi.org/10.1007/s00284-006-0591-7
Liu J, Xing J, Chang T, Ma Z, Liu H (2005) Optimization of nutritional conditions for nattokinase production by Bacillus natto NLSSE using statistical experimental methods. Process Biochem 40(8):2757–2762. https://doi.org/10.1016/j.procbio.2004.12.025
Jo HD, Kwon GH, Park JY, Cha J, Song YS, Kim JH (2011) Cloning and overexpression of aprE3-17 encoding the major fibrinolytic protease of Bacillus licheniformis CH 3-17. Biotechnol Bioproc 16(2):352–359. https://doi.org/10.1007/s12257-010-0328-0
Artemov AV, Samuilov VD (1990) Effect of polyelectrolytes on serine proteinase secretion by Bacillus subtilis. FEBS Lett 262(1):33–35. https://doi.org/10.1016/0014-5793(90)80146-A
Acknowledgements
We express our gratitude to Yan-Chen Man, Chun-Hua Guo and Qian-Rong Xiang for their technical assistance.
Funding
This study was supported by the Mudanjiang Science and Technology Plan Project of China (Nos. G2013n0012 and Z2013n018).
Author information
Authors and Affiliations
Contributions
Li-Li Man performed the experiments and contributed significantly to the analysis and manuscript preparation. Dian-Jun Xiang conceived and designed the experiments. Chun-Lan Zhang performed the data analyses.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Man, LL., Xiang, DJ. & Zhang, CL. Strain Screening from Traditional Fermented Soybean Foods and Induction of Nattokinase Production in Bacillus subtilis MX-6. Probiotics & Antimicro. Prot. 11, 283–294 (2019). https://doi.org/10.1007/s12602-017-9382-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-017-9382-7