Abstract
Wastewater-based epidemiology has been recognized as a tool to monitor the progress of COVID-19 pandemic worldwide. The study presented herein aimed at quantitating the SARS-CoV-2 RNA in the wastewaters, predicting the number of infected individuals in the catchment areas, and correlating it with the clinically reported COVID-19 cases. Wastewater samples (n = 162) from different treatment stages were collected from three wastewater treatment plants (WWTPs) from Mumbai city during the 2nd surge of COVID-19 (April 2021 to June 2021). SARS-CoV-2 causing COVID-19, was detected in 76.2% and 4.8% of raw and secondary treated (n = 63 each) wastewater samples respectively while all tertiary treated samples (n = 36) were negative. The quantity of SARS-CoV-2 RNA determined as gene copies/100 mL varied among all the three WWTPs under study. The gene copy numbers thus obtained were further used to estimate the number of infected individuals within the population served by these WWTPs using two published methods. A positive correlation (p < 0.05) was observed between the estimated number of infected individuals and clinically confirmed COVID-19 cases reported during the sampling period in two WWTPs. Predicted infected individuals calculated in this study were 100 times higher than the reported COVID-19 cases in all the WWTPs assessed. The study findings demonstrated that the present wastewater treatment technologies at the three WWTPs studied were adequate to remove the virus. However, SARS-CoV-2 genome surveillance with emphasis on monitoring its variants should be implemented as a routine practice to prepare for any future surge in infections.
Similar content being viewed by others
Introduction
Coronavirus disease-2019 (COVID-19) is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) belonging to the species SARS-related coronavirus in the subgenus Sarbecovirus of the family Coronaviridae containing enveloped viruses with positive sense single stranded RNA (Kitajima et al., 2020). COVID-19 was first detected in Wuhan, China in December 2019. This disease was later reported in 114 countries and therefore the World Health Organization declared COVID-19 as a pandemic on 11th March 2020 (Medema et al., 2020). As of 29th March 2023, the disease has affected 761,402,282 individuals and 6,887,000 deaths were reported worldwide (WHO-COVID-19 Dashboard accessed at https://covid19.who.int/ on 5th April 2023). In India, 4,47,29,284 confirmed cases and 5,30,901 deaths have been reported till 5th April 2023. (Government of India COVID-19 Dashboard accessed at https://www.mygov.in/covid-19 on 5th April 2023). In India, the first case of COVID -19 was detected in Thrissur, state Kerala on 27th January 2020 while in Mumbai, state Maharashtra, the first case was identified on 11th March, 2020 (https://www.hindustantimes.com/cities/mumbai-news/mumbais-first-covid-patients-we-have-let-it-stay-under-the-carpet-101614797073559.html accessed on 24.11.2022). Individuals infected with COVID-19 infection (symptomatic or asymptomatic) shed SARS-CoV-2 RNA in feces (at a concentration up to 107 copies/g), which is further disposed of in wastewater and may increase the likelihood of fecal–oral transmission (Ahmed et al., 2020; Hemalatha et al., 2021; Kitajima et al., 2020; Pandey et al., 2021).
Surveillance of virus transmission within the population using environmental samples has been established for well-known poliovirus as well as newly emerging Aichi virus (Arora et al., 2020; Hemalatha et al., 2021; Sharma et al., 2021). This type of monitoring is often referred to as Wastewater-Based Epidemiology (WBE) and has also been recognized as a promising surveillance tools for several enteric viruses such as Norovirus, Hepatitis A and E virus, Adenovirus, Rotavirus, and Influenza A (H1N1) and is currently implemented around the globe for SARS-CoV-2 (Arora et al., 2020). WBE has been proven to be effective in monitoring the viral load in the wastewater catchment area, which may provide unambiguous predictions of future outbreaks. It may also aid in uncovering the ground reality of COVID-19 cases (including asymptomatic cases) by examining a larger population as opposed only clinically reported cases in the area (Hata et al., 2020a, 2020b; Srivastava et al., 2021a, 2021b). Various WBE studies for monitoring of SARS-CoV-2 RNA in wastewater have been conducted worldwide in countries such as Australia (Ahmed et al., 2020), Japan (Hata et al., 2020a, 2020b), Netherlands (Medema et al., 2020), Pakistan (Sharif et al., 2020), France (Wurtzer et al., 2020), Spain (Randazzo et al., 2020) and USA (Peccia et al., 2020; Weidhaas et al., 2021).
Several studies have been reported from Indian cities viz. Gandhinagar (Kumar et al., 2021a), Ahmedabad (Joshi et al., 2021; Kumar et al., 2021b) Vadodara (Srivastava et al., 2021b), Bengaluru (Lamba et al., 2023) focusing on estimating the concentration of SARS-CoV-2 in influent wastewater samples received by the wastewater treatment plants in the respective city. Researchers have also used the viral copy number of SARS-CoV-2 data obtained from the wastewater to predict COVID-19 infected individuals (or active shredders) within the community served by the WWTP (Chakraborty et al., 2020; Hemalatha et al., 2021; Lamba et al., 2023). Brian et al., 2022 has summarized India, along with Australia, Japan and the United States in the list of growing number of countries to undertake WBE studies. As SARS-CoV-2 has been widely detected in the influent wastewater samples, the data on efficacy of treatment system to remove the SARS-Cov-2 is very limited. In India, such evaluation has been undertaken only in Jaipur (Arora et al., 2020), Chennai (Chakraborty et al., 2020) and Hyderabad (Hemalatha et al., 2021).
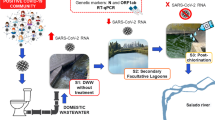
Mumbai is one of the largest and densely populated metropolitan cities in the western coast of India with 6.5 million living in the urban slums (Government of India 2011). The city has recorded 2nd highest number of COVID-19 positive cases (11,57,747) with 19,747 deaths as of 5th April 2023 (Municipal Corporation of Greater Mumbai- accessed at https://stopcoronavirus.mcgm.gov.in/assets/docs/Dashboard.pdf on 5th April 2023). During the initial pandemic phase, one study carried out by Sharma et al. (2021) has reported the presence of SARS-CoV-2 RNA in Mumbai city’s wastewater. In this study, wastewater samples from 5 large open drain and sewage pumping stations between February, March and May 2020 were assessed for the presence of SARS-CoV-2. The city generates 2190 MLD wastewater which is distributed among eight wastewater treatment plants (WWTPs) located across the city. However, each WWTP processes the wastewater up to different stages also using different technologies. viz. WWTP located in Colaba and Charkop are the only two WWTPs carrying out tertiary treatment (chlorination and ozonation, respectively). Bhandup and Versova WWTPs treat the wastewater up to the secondary stage while Bandra, Malad, Ghatkopar and Worli WWTPs treat it up to the primary stage. Of these, 3 WWTPs (Colaba, Charkop and Bhandup) covering south, north and central zone of the city and representing 5,87,930 populations (Table 1), were chosen in the present study.
The present study was undertaken during the 2nd surge of the disease, by when the virus detection systems were also standardized. The objective of the present study was to detect and quantify SARS-CoV-2 RNA in the wastewaters of Mumbai city at various stages of the wastewater treatment process to evaluate treatment systems potential to inactivate/eliminate the SARS-CoV-2 virus from the final effluent. Social distancing/preventive measures in compact places of Mumbai city particularly in slums was a big challenge for the municipal authority, therefore the study also aimed to establish the correlation between the SARS-CoV-2 copy number data obtained from the wastewater with the clinically reported cases from the respective WWTP wards.
Materials and Methods
Sampling Sites and Sample Collection
Raw (influent), secondary and tertiary treated wastewater (effluent) samples were collected by grab sampling technique from three WWTPs. Bhandup WWTP (WWTP-Z3), located in zone-3 under the ‘S’ administrative ward of Mumbai Municipal Corporation of Greater Mumbai (MCGM). Colaba WWTP (WWTP-Z1), situated in zone1 (city center) is under 'A' ward of MCGM and Charkop WWTP (WWTP-Z5) in zone 5 of Mumbai city under ‘R/South’ ward of MCGM (Fig. 1). Technical details of the wastewater treatment at the three WWTPs are compiled in Table 1. A total of 162 wastewater samples (1 L) were collected in a sterile polypropylene (PP) containers which comprised paired raw/influent and secondary treated wastewater samples collected from WWTP-Z3 (n = 27), WWTP-Z5 (n = 22) and WWTP-Z1 (n = 14) WWTP, respectively. Tertiary treated wastewater were also collected from WWTP-Z5 (n = 22) and WWTP-Z1 (n = 14) WWTP, respectively. All samples were collected during April 2021 to June 2021 on weekdays (Monday to Thursday/Friday) between 9:00 AM-10:00 AM. This sampling period covered the 2nd wave of COVID-19 pandemic in Mumbai city. However, due to the lockdown and restricted travel, there was a gap of 2 weeks during the sampling.
Sample Processing and SARS-CoV-2 Concentration
Samples were processed within 3 h post collection on the same day. Standardized PEG precipitation method (Michael-kordatou et al., 2020) was used to concentrate SARS-CoV-2 in wastewater samples by taking 100 mL of wastewater sample, centrifuged at 4000×g for 20 min (Eppendorf Centrifuge 5804R) to remove the coarse debris. The supernatant was then transferred into sterile screw cap glass bottle containing 8% (w/vol) polyethylene glycol (PEG) (Sigma Aldrich, USA) and 0.2 M (w/v) sodium chloride (Sigma Aldrich, USA). Bacteriophage Phi6 (DSM 21518), spiked at 5.5. × 105 pfu/100 mL of sewage was used as an internal process control to assess recovery of enveloped RNA viruses from wastewater samples (Kitajima et al., 2020). The samples were transferred to an orbital shaker incubator (Trishul Equipments India) adjusted at 4 °C and 130 RPM for 2 h and then subsequently held at 2–8 °C for virus precipitation (Flood et al., 2021). The virus precipitate was pelleted out by centrifugation at 10,000×g for 30 min at 4 °C and the pellet was resuspended in 2 mL PBS (Themofisher, USA). The virus concentrates were stored at − 80 °C. RNA extraction and the subsequent qPCR assay was performed within 72 h.
Qualitative Detection of SARS-CoV-2
For qualitative detection of SARS-CoV-2, 450 µL of virus concentrate was mixed with 50 µL of uninfected human plasma (NHP) and was used for RNA extraction. RNA was extracted using the Roche HiPure system viral nucleic acid kit (Roche, USA) as per the manufacturer’s instructions. The RNA was eluted in 75 µL of elution buffer and used for the detection of SARS-CoV-2 using TRUPCR SARS-CoV-2 RT- qPCR kit (V-3.2) (3B BlackBio Biotech India Ltd., India) in StepOne™ Instrument (Applied Biosystems, USA). The procedure included addition of 10 µL of extracted RNA to 15 µL of total reaction mix which contained master mix (10 µL), enzyme mix (0.35 µL) and primer–probe mix (4.65 µL). The RT qPCR program set up was 1 cycle each at 50 °C/15 min and 95 °C/05 min followed by 38 cycles of 95 °C/05 s, 60 °C/40 s (included dye acquisition) and 72 °C/15 s. Each run included negative and positive control provided with the kit.
Quantification of SARS-CoV-2 in Wastewater Samples Using qRT-PCR
Wastewater samples tested positive qualitatively were subjected to quantitative reverse transcriptase PCR assay to determine the RNA copies of SARS-CoV-2. RNA extracted from 300 µL of virus concentrate using the HiPure system viral nucleic acid kit (Roche, USA) was eluted in 50 µL of nuclease free water. Specific gene targets viz. N1 (Nucleocapsid), ORF1b-nsp14 (located in Open Reading Frame 1b) and RdRp (RNA dependent RNA polymerase) were selected for quantification of SARS-CoV-2 RNA. Additionally, Phi6 used as an internal process control was also quantified. The sequences of primers-probes used are given in Table 2. 5 µL of RNA template was used in a total reaction volume of 25 µL containing SuperScript™ III RT/Platinum Taq Mix (0.5 µL), 2X Reaction mix (12.5 µL), 10 μm forward and reverse primer (0.5 µL), 10 μM fluorogenic probe 0.25 µL. qRT-PCR cycling program was set as 1 cycle of each 50 °C/15 min (cDNA synthesis) and 95 °C/2 min followed by 40 cycles each of 95 °C/15 s, 60 °C/30 s. Quantitative PCR was carried out using SuperScript™ III Platinum™ One-Step qRT-PCR Kit (Invitrogen, USA) in Applied Biosystems StepOnePlus™ Instrument. Gene copies/reaction obtained from the assay was used to back calculate gene copies/100 mL of wastewater samples. SARS-CoV-2 quantitative RNA reference standard (ATCC 3276SD) was used as a standard copy control.
COVID-19 Reported Cases in WWTP Catchment Zone and Number of Infectious Individuals in the Population Served by 3 WWTP
Data on the number of daily reported COVID-19 cases during the study (April to June 2021) was provided by the public health officers of the respective MCGM administrative wards. Information related to the area served by the WWTP was obtained from the respective ward offices. Number of infected individuals among the population served by each WWTP was estimated using 2 different methods that have been previously reported (Ahmed et al., 2020; Hemalatha et al., 2021).
The equations used for calculation are mentioned below:
-
Method 1 (Eq. 1): (Ahmed et al., 2020)
$${\text{Persons\; Infected}} = \frac{{\left( {\frac{{\text{RNA\; copies}}}{{\text{liter\; wastewater}}}} \right) \times \left( {\frac{{\text{liters\; wastewater}}}{{{\text{day}}}}} \right)}}{{\left( {\frac{{\text{g\; feces}}}{{\text{person-day}}}} \right) \times \left( {\frac{{\text{RNA\; copies}}}{{\text{g\; feces}}}} \right)}}$$(1)For g feces, 128 g was used for feces excreted/person/day and it has been estimated that, positive individual shed 107 RNA copies/g of feces (Ahmed et al., 2020).
-
Method 2 (Eq. 2): (Hemalatha et al., 2021)
$${\text{Persons\; Infected }} = \frac{{{\text{No.\;of \;RNA \;copies\; per\; liter\; of\; wastewater}}}}{{\text{Contribution\; of\; RNA\; copies\; per\; person\; to\; total\; sewage\; water}}}$$(2)107 RNA copies/g of feces was multiplied with 120 mL of volume of faeces excreted by human (considering density of feces as 1.07 g/mL) and total wastewaters (L) received at WWTP (Hemalatha et al., 2021).
Data Analysis
The data obtained from qualitative and quantitative estimation of SARS-CoV-2 RNA as well as Phi6 was transferred and analyzed in Microsoft Excel spreadsheet. IBM SPSS version 23.0 was used for data visualization and statistical analysis. Based on the Phi6 and the SARS-CoV-2 standard copy control, sample limit of detection and assay limit of detection was determined. The copy controls were used to calculate the gene copies of Phi6 and SARS-CoV-2 per reaction in each of the samples and was subsequently back calculated to gene copies/ 100 mL of sewage. Correlational analysis was performed to determine correlation of average SARS-CoV-2 GC/100 mL quantities with physico-chemical parameters, reported COVID-19 cases, and infectious individuals in the population calculated using three methods and p value < 0.05 was considered as a statistically significant correlation. Qualitatively positive SARS-CoV-2 samples that were undetected for a particular gene were assigned the sample limit of detection for correlational analysis.
Results
Qualitative Detection of SARS-CoV-2 in Wastewater Samples
To assess the prevalence of SARS-CoV-2 in the wastewater samples, TRUPCR SARS-CoV-2 RT- qPCR kits (V-3.2) were used. The dual gene targets (RdRp + N) used for qualitative detection of SARS-CoV-2 was detected in 76.2% (48 out of 63) of raw wastewater samples (mean Ct. value 31.5 ± 3.2). Of the 63 secondary treated wastewater samples, only three (4.8%) from WWTP-Z3 (Mean Ct. value 34.3 ± 0.5) were positive for SARS-CoV-2. The remaining 60 secondary treated samples as well as all the tertiary treated samples (n = 36) collected in this study were negative for SARS-CoV-2.
Quantification of Phi6 and SARS-CoV-2 in Wastewater Samples
Recovery of Internal Process Control Phi6
Phi6, an enveloped RNA bacteriophage, has been used as a surrogate for viral respiratory pathogens such as SARS-CoV-2 in wastewater analysis because of its resemblance to those viral respiratory pathogens and absence in urban wastewater (Fedorenko et al., 2020; Kitajima et al., 2020). Thus, an internal process control Phi6 was added to all the samples including the negative control to assess the recovery of enveloped viruses using RT-qPCR. Phi6 was detected in all the spiked samples with the mean cycle threshold (Ct) value of 29.7 ± 3.2. (Mean ± S.D.) and the mean percentage recovery of 33.9 ± 29.9% (Mean ± S.D.).
Quantification of SARS-CoV-2 in Wastewater
The wastewater samples positive for SARS-CoV-2 RNA by qualitative method were subjected to quantitative PCR for 3 gene targets viz. N1, ORF1b-nsp14 and RdRp. The performance efficiency of RT-qPCR assay for SARS-CoV-2 and Phi6 gene targets have been summarized in Table 3. The Ct values and the calculated mean gene copies/100 mL of wastewater samples are provided in Table 4. The mean N1, ORF1b-nsp14 and RdRp genes copies in WWTP-Z3 and WWTP-Z5 were in the range of 105 GC/100 mL (except for N1 and RdRp gene quantified in WWTP-Z3 were 104 GC/100 mL) and were higher by 52.6% and 85.9% of gene copies, respectively as compared to WWTP-Z1 (104 GC/100 mL). However, amongst the three WWTPs, SARS-CoV-2 gene copies obtained were highest in WWTP-Z5. These observed variations in the quantities of three genes in the raw wastewaters across the three WWTPs is represented using the box-whisker plot (Fig. 2).
Reported Cases of COVID-19 During Wastewater Surveillance Period and Correlational Analysis with SARS-CoV-2
The study period was in the middle of the 2nd surge of COVID-19 cases in Mumbai city, the clinically confirmed COVID-19 case load was retrieved from the MCGM wards. It was observed that the number of daily reported cases from WWTP-Z3 (S ward) and WWTP-Z5 (R/South ward) decreased from 430 and 519 cases/day in April 2021 to 43 and 48 cases/day in the month of June 2021, respectively. Similar trend was also observed in WWTP-Z1 (A ward) wherein during the study period (May and June 2021), COVID-19 cases decreased from 30 cases/day in May 2021 to a mere five cases/day by June 2021. These observations were in-line with SARS-CoV-2 gene copy numbers observed in the wastewater samples of the respective wards (Fig. 3). A positive trend was observed between the weekly average SARS-CoV-2 gene copy number in the wastewater and the cumulative weekly reported COVID-19 cases in all the three WWTPs. A statistically significant strong positive correlation was obtained with WWTP-Z3 (0.863, p = 0.027) and WWTP-Z5 (0.859, p = 0.028) WWTP (Table 5). WWTP-Z1 showed moderate positive correlation (− 0.31, p = 0.69) however, this association was not statistically significant.
Week-wise trend of mean SARS-CoV-2 gene copies estimated in raw wastewater versus reported COVID-19 cases in the WWTPs served area. A WWTP-Z1 (Colaba), B WWTP-Z3 (Bhandup) and C WWTP-Z5 (Charkop). Bars represent weekly mean SARS-CoV-2 gene copies/100 mL of sewage and Line represents the weekly mean COVID-19 cases for the ward area served by the WWTP
Number of Infected Individuals and Correlation with COVID-19 Reported Cases
Two methods were used to estimate the number of infected individuals among the population served by WWTP based on the SARS-CoV-2 gene copy number obtained from the wastewater samples (Ahmed et al., 2020; Hemalatha et al., 2021). It was observed that the number of infected individuals estimated by the method 1 and method 2 were similar and in the range of 104–105 (Fig. 4). Correlational analysis of reported cases with the estimated number of infected individuals are shown in Table 5. The predicted number of infected individuals using the two methods followed a decreasing trend similar to the reported COVID-19 cases and a statistically significant correlation was obtained with data from WWTP-Z3 (r = 0.863; p = 0.027) and WWTP-Z5 (r = 0.859; p = 0.028) but not with WWTP-Z1 (r = − 0.31; p = 0.69).
Reported and Predicted COVID 19 infected cases across 3 WWTPs. The figure represents mean of week-wise COVID-19 reported cases and predicted infected individuals using Ahmed et al. (2020) and Hemalatha et al. (2021) methods using Wastewater-based Epidemiology approach. The correlation between reported and predicted COVID-19 cases was significant (p < 0.05) among WWTP-Z3 and Z5 and vice-versa
Discussion
The present study conducted in the city of Mumbai, India aimed to provide an in-depth analysis of the prevalence of SARS-CoV-2 RNA viral copies in the wastewater and its persistence through the treatment process during the 2nd surge of the COVID-19 pandemic. The study aimed to evaluate the efficiency of the treatment systems (followed at three WWTPs) to eliminate the SARS-CoV-2 virus from the wastewater. The obtained SARS-CoV-2 RNA copy data was used to establish correlation between the SARS-CoV-2 RNA copies in the untreated wastewater and the reported clinical infections from the areas served by the WWTP.
During sample processing, bacteriophage phi-6 was incorporated in all the samples as a process control to assess virus recovery and the samples were processed using the PEG/NaCl to obtain the virus concentrates (Michael-kordatou et al., 2020). The bacteriophage phi-6 was selected as a process control due to its similarity to enveloped RNA viruses and its substantiated use as a surrogate for various respiratory viruses such as Ebola virus, influenza virus, coronaviruses (SARS-1) and recently, SARS-CoV-2 (Fedorenko et al., 2020; Kitajima et al., 2020; Whitworth et al., 2020). The Phi6 recoveries observed in the study (22–36%) were similar to values reported by Flood et al. (2021) using similar method. Many researchers have higher recoveries of phi6 (56% and 54%) using ultrafiltration and adsorption-elution methods for virus concentration respectively (Sherchan et al., 2020). The recovery of the phi6 using PEG/NaCl method in the present study is similar to that reported by Flood et al. (2021) (22–36%). The satisfactory virus recovery along with the cost-effectiveness of PEG/NaCl method substantiates its use particularly in the resource limited settings.
The presence of SARs-CoV-2 in the wastewater samples was qualitatively detected using TRUPCR SARS-CoV-2 RT- qPCR kit (V-3.2) (3B BlackBio Biotech India Ltd, India) which is an Indian Council of Medical Research (ICMR) approved kit for qualitative real-time PCR kit. The kit is very sensitive with a limit of detection of 6 copies/ µL of reaction. This high sensitivity is attributed to the use of dual probes with single dye against the RdRp gene and N gene for the detection of SARS-CoV-2, thus preventing any ambiguity resulting from mutations in any one of the target region. It was observed that the SARS-CoV-2 RNA was detected in majority of the untreated wastewater samples (76.2%, n = 48/63) indicating prevalence of COVID-19 disease in the population served by the three WWTPs under study. The SARS-CoV-2 RNA was undetected in most of the secondary and all of the tertiary treated in WWTPs Z1 and WWTP Z5.
Z1 and Z5 WWTP use the Rotating Media Bio Reactor (RMBR) and Sequential Batch Reactor (SBR) technology for secondary treatment, respectively. These technologies aid in efficient separation of the generated sludge from the wastewater which is also known to drastically reduce bacterial and viral load from the wastewater (Bhave et al., 2020), thus no viral RNA was detected post-secondary treatment herein. Arora et al. (2020) also reported absence of SARS-CoV-2 in the treated wastewater samples processed through similar methods. However, it would be worthwhile to check the presence of SARS-CoV-2 RNA or the live virus in the sludge generated from the WWTPs that subsequently ends up in landfills or in gardens. The lagoon-based secondary treatment followed at WWTP-Z3 was able to reduce the SARS-CoV-2 sample positivity by 95.2%; with 3 samples out of 27 from this WWTP testing positive for SARS-CoV-2 RNA. This indicates the limitation of this method of secondary treatment in the virus elimination. Similar findings have been reported from a Spain-based study using similar treatment technology (Randazzo et al., 2020), wherein 11% of the secondary treated samples (n = 18, from 2 different WWTPs) were positive for SARS-CoV-2. Inability of this treatment procedure, may be due to the large area of the lagoon resulting in inefficient uniform aeration. However, this assumption needs to be validated.
Post qualitative assessment, SARS-COV-2 viral copies in the wastewater were estimated using three targets genes of SARS-CoV-2 namely N1, ORF1b-nsp14 and RdRp gene Selection of these genes was based on recommendations by national and international bodies. N1 gene was recommended by Center for Disease Control (CDC), Atlanta, USA and has also been commonly used in several wastewater-based studies worldwide (Haramoto et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Sherchan et al., 2020). The use of ORF1b-nsp14 and RdRp was as per genes targets reported for clinical and environmental studies by several researchers (Corman et al., 2020; Kitajima et al., 2020; Rosa et al., 2020) however, the RT-qPCR efficiencies have not been discussed and as per the recommendation by ICMR, Delhi, India),. for this surveillance (https://www.icmr.gov.in/pdf/covid/labs/2_SOP_for_Confirmatory_Assay_for_2019_nCoV.pdf accessed on 19th June, 2022).
We observed that the PCR efficiency for Orf-1b-nsp-14 and RdRp was in the range of 71–75%) while that of N1 gene was 87%. Considering that the same samples were used for all the three gene targets the used of ORF1-b-nsp-14 and RdRp should be studied further for its suitability in wastewater-based surveillance. Quantitative data on SARS-CoV-2 wastewater surveillance studies have been reported from other parts of India during different phases of the pandemics with variable gene copy numbers. Chakraborty et al., 2020 reported 104–105 GC/L of SARS-CoV-2 (N1 and N2 genes) in 4 WWTPs from Chennai city during September 2020. Srivastava et al. (2021a, 2021b) reported lower gene copies (102–103 GC/L) of SARS-CoV-2 (N, ORF and S gene) in untreated wastewaters of Gandhinagar, Ahmedabad and Vadodara cities in India during November 2020. Nag et al. (2022), studied 11 WWTP’s across Jaipur city during Feb to Jun 2021 and obtained SARS-CoV-2 gene (E, RdRp and N) concentrations in the range of 1 × 104 to 1 × 106 gene copies/L of wastewater.
From Mumbai city wastewaters, SARS-CoV-2 virus was studied in the early phase of the pandemic (Feb 2020 to May 2020). The study was qualitative, using E and RdRp gene sampled largely from 5 open drains and one sewage pumping station (Sharma et al., 2021) This study, thus was designed to understand the quantitative data of a large metropolitan city, also the virus fate in waste water treatment and finally correlating the quantitative data with clinical cases, contributing to WBE. The study conducted here estimated the SARS-CoV-2 viral copy numbers in the wastewater was carried out between April–June 2021. It was observed that the viral copy numbers were higher (104–105 GC/100 mL) in untreated wastewater samples and was tenfold higher in WWTP-Z5 (105 GC/100 mL) as compared to WWTP-Z1and WWTP-Z3 (104 GC/100 mL). This variation between the SARS-CoV-2 copy number observed in the WWTPs could largely be attributed to number of prevalent COVID-19 cases among the population, the population density and the amount of wastewater generated by the community served by the WWTP. Even though WWTP-Z5 had the lowest volume of wastewater treated per day amongst the three WWTPs, this WWTP mainly serves only the residential area which may contribute to higher SARS-CoV-2 viral load as compared to WWTP-Z3 and WWTP-Z5 where the chance of wastewater getting diluted due to industrial activities cannot be excluded. Similar observations were also reported by Kumar et al. (2021a) where N, ORF and S genes were used for the study. The three secondary treated samples from WWTP-Z3 that were qualitatively positive for SARS-CoV-2 were also positive by quantitative tests for at least two genes. However, there was a 1.38 Log10 reduction in the SARS-CoV-2 RNA copies/100 mL when compared to untreated /raw wastewater.
In the present study, a statistically significant strong positive correlation was observed between the weekly mean SARS-CoV-2 RNA copies from wastewater and weekly mean confirmed COVID-19 cases reported from the catchment areas of two WWTP’s (Z3 and Z5). Similar correlations were also observed by Kumar et al. (2021a, 2021b) and Srivastava et al. (2021a, 2021b) in studies conducted across various cities (Gandhinagar, Vadodara and Ahmedabad) across Gujarat. However, such correlation was not observed in WWTP-Z1 wherein the sampling was initiated during the late stage of the 2nd surge of COVID-19 infection where the reported case load from this municipal ward was also very low. Thus, this inconsistency may have been one of the contributing factor. Additionally, using the weekly mean SARS-CoV-2 gene copy number data from the wastewater, the number of active shedders was estimated by two published methods (Ahmed et al., 2020; Hemalatha et al., 2021). Although a positive trend (tenfold decrease) was observed in the estimated SARS-CoV-2 gene copies and reported COVID-19 cases across all 3 WWTPs (Table 5), the predicted caseloads were 100 times higher (median value) than clinically confirmed COVID-19 cases (Table 6). This difference may be a result of lag in the active reporting of cases, the prevalence of asymptomatic infections within the community as well as the enrichment of SARS-CoV-2 copies due to grab sampling method during the peak hours of the wastewater flow. The latter may become a contributing factor as the concentration of SARS-CoV-2 in the grab samples may vary at different time points.
It is important to highlight that the models used in this study were based on the available models developed during the early phase of the pandemic which may be crude as compared to some of the current models developed. Recent studies (Oh et al., 2022; Zhan et al., 2022) have incorporated RT-qPCR data normalization using the PMMoV (Pepper Mild Mottle Virus) which is naturally present in the wastewater (contributed as part of human faeces) which may be more robust in estimating the depth of infection in the community.
Though the study was designed using information, methods and data available during the 1st quarter of 2021, there were also a few limitations that were beyond our control while executing the project. Although simultaneous sampling was initially envisaged in this study, strict lockdown imposed by Maharashtra State due to emergence of SARS-CoV-2 Delta variant (2nd COVID-19 wave in 2021) hampered continuous sampling from the three WWTPs. One cannot deny that these changes may partly affect the correlation co-efficient observed in this study. Furthermore, grab sampling method was used assuming the enriched samples at peak hours between 9 and 10 am over composite sampling, to avoid dilution of SARS-CoV-2 in composite samples.
To conclude, this study was undertaken to assess the presence of SARS-CoV-2 in the wastewater samples in three WWTPs of Mumbai city during the 2nd surge of COVID-19. The data obtained suggests that 1) PEG/NaCl method was relatively effective to concentrate the virus from the wastewater and can be used in resource limited settings. 2) SARS-CoV-2 RNA was efficiently eliminated during wastewater treatment from all the three WWTPs including the one with just a secondary treatment facility. 3) Although qualitative method can be used for routine screening of the wastewaters for SARS-CoV-2 RNA while quantitative data provides the number of active shedders who are already infected in the geographical area and hence giving an estimate of cases to manage any surge in infections in the future 4) The study also highlights the importance of upgrading the WWTPs up to tertiary treatment for reduction of the virus to non-detectable levels as observed in the two WWTPs equipped with tertiary treatment (WWTP-Z5 and WWTP-Z1) and 5) Continued monitoring of SARS-CoV-2 including its variants in wastewater is suggested for early warning system.
References
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., Brien, J. W. O., Choi, P. M., Kitajima, M., Simpson, S. L., Li, J., Tscharke, B., Verhagen, R., Smith, W. J. M., Zaugg, J., Dierens, L., Hugenholtz, P., Thomas, K. V., & Mueller, J. F. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia : A proof of concept for the wastewater surveillance of COVID- 19 in the community. Science of the Total Environment, 728, 138764. https://doi.org/10.1016/j.scitotenv.2020.138764
Arora, S., Nag, A., Sethi, J., Rajvanshi, J., Saxena, S., Shrivastava, S. K., & Gupta, A. B. (2020). Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. Water Science and Technology, 82(12), 2823–2836. https://doi.org/10.2166/wst.2020.540
Bhave, P. P., Naik, S., & Salunkhe, S. D. (2020). Performance evaluation of wastewater treatment plant. Water Conservation Science and Engineering, 5(1), 23–9.
Brian, I., Manuzzi, A., Dalla Rovere, G., Giussani, E., Palumbo, E., Fusaro, A., Bonfante, F., Bortolami, A., Quaranta, E. G., Monne, I., Patarnello, T., Bargelloni, L., Terregino, C., Holmes, E. C., Todesco, G., Sorrentino, F., Berton, A., Badetti, C., Carrer, C., … Panzarin, V. (2022). Molecular Monitoring of SARS-CoV-2 in Different Sewage Plants in Venice and the Implications for Genetic Surveillance. ACS ES and T Water, 2(11), 1953–1963. https://doi.org/10.1021/acsestwater.2c00013
Chakraborty, P., Pasupuleti, M., Shankar, M. R. J., Bharat, G. K., & Krishnasamy, S. (2020). First surveillance of SARS-CoV-2 and organic tracers in community wastewater during post lockdown in Chennai, South India: Methods, occurrence and concurrence. Science of the Total Environment, 778, 146252–54.
Corman, V., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K., Bleicker, T., Brünink, S., Schneider, J., Luisa Schmidt, M., Mulders, D., Haagmans, B. L., van der Veer, B., van den Brink, S., Wijsman, L., Goderski, G., Romette, J.-L., Ellis, J., Zambon, M., … Chantal, R. (2020). Detection of 2019-nCoV by RT-PCR. Eurosurveillance, 25(3), 1–8.
Fedorenko, A., Grinberg, M., Orevi, T., & Kashtan, N. (2020). Survival of the enveloped bacteriophage Phi6 (a surrogate for SARS-CoV-2) in evaporated saliva microdroplets deposited on glass surfaces. Scientific Reports, 6, 1–10. https://doi.org/10.1038/s41598-020-79625-z
Flood, M. T., Souza, N. D., Rose, J. B., & Gim, T. (2021). Methods evaluation for rapid concentration and quantification of SARS-CoV-2 in raw wastewater using droplet digital and quantitative RT-PCR. Food and Environmental Virology, 13(3), 303–315. https://doi.org/10.1007/s12560-021-09488-8
Gendron, L., Verreault, D., Veillette, M., Moineau, S., & Duchaine, C. (2010). Evaluation of filters for the sampling and quantification of RNA phage aerosols. Aerosol Science and Technology, 44(10), 893–901. https://doi.org/10.1080/02786826.2010.501351
Government of India. (2011). Slums in India: A statistical compendium. https://nbo.gov.in/pdf/Slum_in_india_2011_english_book_23_May_12.pdf. Accessed 30 Nov 2022.
Haramoto, E., Malla, B., Thakali, O., & Kitajima, M. (2020). First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Science of the Total Environment, 737, 140405.
Hata, A., Hara-yamamura, H., Meuchi, Y., Imai, S., & Honda, R. (2020a). Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Science of the Total Environment, 758, 143578.
Hata, A., Honda, R., Hara-Yamamura, H., & Meuchi, Y. (2020b). Detection of SARS-CoV-2 in wastewater in Japan by multiple molecular assays-implication for wastewater-based epidemiology (WBE). MedRxiv. https://doi.org/10.1101/2020.06.09.20126417
Hemalatha, M., Kiran, U., Kuncha, S. K., Kopperi, H., Gokulan, C. G., Mohan, S. V., & Mishra, R. K. (2021). Surveillance of SARS-CoV-2 spread using wastewater-based epidemiology: Comprehensive study. Science of the Total Environment, 768, 144704. https://doi.org/10.1016/j.scitotenv.2020.144704
Joshi, M., Kumar, M., Srivastava, V., & Kumar, D. (2021). First detection of SARS-CoV-2 Delta variant (B.1.617.2) in the wastewater of (Ahmedabad), India. MedRxiv. https://doi.org/10.1101/2021.07.07.21260142
Kitajima, M., Ahmed, W., Bibby, K., Carducci, A., Gerba, C. P., Hamilton, K. A., Haramoto, E., & Rose, J. B. (2020). SARS-CoV-2 in wastewater: state of the knowledge and research needs Masaaki. Sci Total Environ, 739, 139076.
Kumar, M., Joshi, M., Patel, A. K., & Joshi, C. G. (2021). Unravelling the early warning capability of wastewater surveillance for COVID-19: A temporal study on SARS-CoV-2 RNA detection and need for the escalation. Environmental Research, 196, 110946. https://doi.org/10.1016/j.envres.2021.110946
Kumar, M., Joshi, M., Shah, A. V., Srivastava, V., & Dave, S. (2021). Wastewater surveillance-based city zonation for effective COVID-19 pandemic preparedness powered by early warning: A perspectives of temporal variations in SARS-CoV-2-RNA in Ahmedabad, India. Science of the Total Environment, 792, 148367. https://doi.org/10.1016/j.scitotenv.2021.148367
La Rosa, G., Iaconelli, M., Mancini, P., Bonanno Ferraro, G., Veneri, C., Bonadonna, L., Lucentini, L., & Suffredini, E. (2020). First detection of SARS-CoV-2 in untreated wastewaters in Italy. Science of the Total Environment, 736, 139652. https://doi.org/10.1016/j.scitotenv.2020.139652
Lamba, S., Ganesan, S., Daroch, N., Paul, K., Joshi, S. G., Sreenivas, D., Nataraj, A., Srikantaiah, V., Mishra, R., Ramakrishnan, U., & Ishtiaq, F. (2023). SARS-CoV-2 infection dynamics and genomic surveillance to detect variants in wastewater—a longitudinal study in Bengaluru, India. The Lancet Regional Health—Southeast Asia, 11, 100151. https://doi.org/10.1016/j.lansea.2023.100151
Medema, G., Heijnen, L., Italiaander, R., & Brouwer, A. (2020). Presence of SARS-coronavirus—2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environmental Science & Technology Letters. https://doi.org/10.1021/acs.estlett.0c00357
Michael-kordatou, I., Karaolia, P., & Fatta-kassinos, D. (2020). Making waves: Wastewater surveillance of SARS-CoV-2 for population-based health management. Journal of Environmental Chemical Engineering, 8(January), 1–24. https://doi.org/10.1016/j.jece.2020.104306
Nag, A., Arora, S., Sinha, V., Meena, E., Sutaria, D., Gupta, A. B., & Medicherla, K. M. (2022). Monitoring of SARS-CoV-2 variants by wastewater-based surveillance as a sustainable and pragmatic approach—a case study of Jaipur (India). Water (Switzerland), 14(3), 1–19. https://doi.org/10.3390/w14030297
Oh, C., Zhou, A., O’Brien, K., Jamal, Y., Wennerdahl, H., Schmidt, A. R., Shisler, J. L., Jutla, A., Keefer, L., Brown, W. M., & Nguyen, T. H. (2022). Application of neighborhood-scale wastewater-based epidemiology in low COVID-19 incidence situations. Science of the Total Environment, 852(June), 158448. https://doi.org/10.1016/j.scitotenv.2022.158448
Pandey, D., Verma, S., Verma, P., Mahanty, B., Dutta, K., Daverey, A., & Arunachalam, K. (2021). SARS-CoV-2 in wastewater: Challenges for developing countries. International Journal of Hygiene and Environmental Health, 231, 113634. https://doi.org/10.1016/j.ijheh.2020.113634
Peccia, J., Zulli, A., Brackney, D. E., Grubaugh, N. D., Kaplan, E. H., Casanovas-Massana, A., Ko, A. I., Malik, A. A., Wang, D., Wang, M., Warren, J. L., Weinberger, D. M., Arnold, W., & Omer, S. B. (2020). Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nature Biotechnology, 38(10), 1164–1167. https://doi.org/10.1038/s41587-020-0684-z
Randazzo, W., Truchadc, P., Cuevas-Ferrando, E., Simón, P., Allende, A., & Sánchez, G. (2020). SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Research, 181, 115942.
Sharif, S., Ikram, A., Khurshid, A., Salman, M., Mehmood, N., Arshad, Y., Ahmad, J., Angez, M., Alam, M. M., Rehman, L., Mujtaba, G., Hussain, J., Ali, J., Akthar, R., Malik, M. W., Baig, Z. I., Rana, M. S., Usman, M., Qasir, M., … Tahir, F. (2020). Detection of SARs-CoV-2 in wastewater, using the existing environmental surveillance network : An epidemiological gateway to an early warning for COVID-19 in communities Introduction. PLoS ONE. https://doi.org/10.1371/journal.pone.0249568.
Sharma, D. K., Nalavade, U. P., Kalgutkar, K., Gupta, N., & Deshpande, J. M. (2021). SARS-CoV-2 detection in sewage samples: Standardization of method & preliminary observations. Indian Journal of Medical Research, 153, 159–165. https://doi.org/10.4103/ijmr.IJMR_3541_20
Sherchan, S. P., Shahin, S., Ward, L. M., Tandukar, S., Aw, T. G., Schmitz, B., Ahmed, W., & Kitajima, M. (2020). First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Science of the Total Environment, 743, 140621. https://doi.org/10.1016/j.scitotenv.2020.140621
Srivastava, V., Gupta, S., Patel, A. K., Joshi, M., & Kumar, M. (2021b). Reflections of COVID-19 cases in the wastewater loading of SARS-CoV-2 RNA: A case of three major cities of Gujarat, India. In Case studies in chemical and environmental engineering (Vol. 4). Elsevier. https://doi.org/10.1016/j.cscee.2021.100115
Srivastava, V., Gupta, S., Patel, A. K., Joshi, M., & Kumar, M. (2021a). Reflections of COVID-19 cases in the wastewater loading of SARS-CoV-2 RNA: A case of three major cities of Gujarat, India. Case Studies in Chemical and Environmental Engineering, 4(April), 4–9. https://doi.org/10.1016/j.cscee.2021.100115
Weidhaas, J., Aanderud, Z. T., Roper, D. K., VanDerslice, J., Gaddis, E. B., Ostermiller, J., Hoffman, K., Jamal, R., Heck, P., Zhang, Y., Torgersen, K., Laan, J. Vander., & LaCross, N. (2021). Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Science of the Total Environment, 775, 145790. https://doi.org/10.1016/j.scitotenv.2021.145790
Whitworth, C., Mu, Y., Houston, H., Martinez-Smith, M., Noble-Wang, J., Coulliette-Salmond, A., & Rose, L. (2020). Persistence of bacteriophage Phi 6 on porous and nonporous surfaces and the potential for its use as an Ebola virus surrogate. Applied Environmental in Microbiology, 86(17), 1–11.
Wurtzer, S., Marechal, V., Mouchel, J. M., Maday, Y., Teyssou, R., Richard, E., Almayrac, J. L., & Moulin, L. (2020). Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris in wastewater, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance Weekly, 25(50), 2000776.
Zhan, Q., Babler, K. M., Sharkey, M. E., Amirali, A., Beaver, C. C., Boone, M. M., Comerford, S., Cooper, D., Cortizas, E. M., Currall, B. B., Foox, J., Grills, G. S., Kobetz, E., Kumar, N., Laine, J., Lamar, W. E., Mantero, A. M. A., Mason, C. E., Reding, B. D., … Solo-Gabriele, H. M. (2022). Relationships between SARS-CoV-2 in wastewater and COVID-19 clinical cases and hospitalizations, with and without normalization against indicators of human Waste. ACS Environmental Science & Technology Water, 2(11), 1992–2003. https://doi.org/10.1021/acsestwater.2c00045
Acknowledgements
This work is funded by the Indo-US Science and Technology Forum (IUSSTF) [Ref No: IUSSTF/VN-COVID/081/2020]. We sincerely thank the authorities of Municipal Corporation of Greater Mumbai (MCGM) as well as the technical team working at all the WWTPs for their kind assistance. We thank Mr. Mayur Shelar and Mr. Shivam Nikam for helping with sample collection and transportation. We appreciate the assistance of all the BRC staff for their co-operation in this work.
Author information
Authors and Affiliations
Contributions
ND, ZB, JR and SS conceptualized the study, NDS and JR trained the BRC staff in sample processing and data analysis, DD, HW and SM coordinated sample collection, HW and SM carried out sample processing, HW, DD and ND analyzed the data, HW wrote the draft manuscript and DD edited the draft manuscript, all authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wani, H., Menon, S., Desai, D. et al. Wastewater-Based Epidemiology of SARS-CoV-2: Assessing Prevalence and Correlation with Clinical Cases. Food Environ Virol 15, 131–143 (2023). https://doi.org/10.1007/s12560-023-09555-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-023-09555-2