Abstract
The COVID-19 pandemic affected human life at every level. In this study, we analyzed genetic markers (N and ORF1ab, RNA genes) of SARS-CoV-2 in domestic wastewaters (DWW) in San Justo City (Santa Fe, Argentina), using reverse transcription-quantitative real-time PCR. Out of the 30 analyzed samples, 30% were positive for SARS-CoV-2 RNA. Of the total positive samples, 77% correspond to untreated DWW, 23% to pre-chlorination, and no SARS-CoV-2 RNA was registered at the post-chlorination sampling site. The viral loads of N and OFR1ab genes decreased significantly along the treatment process, and the increase in the number of viral copies of the N gene could anticipate, by 6 days, the number of clinical cases in the population. The concentration of chlorine recommended by the WHO (≥ 0.5 mg L−1 after at least 30 min of contact time at pH 8.0) successfully removed SARS-CoV-2 RNA from DWW. The efficiency of wastewater-based epidemiology (WBE) confirms the need to control and increase DWW treatment systems on a regional and global scale. This work could contribute to building a network for WBE to monitor SARS-CoV-2 in wastewaters during the pandemic waves and the epidemic remission phase.
Graphical abstract

Similar content being viewed by others
1 Introduction
An efficient treatment of DWW has become a great challenge with the outbreak of the COVID-19 pandemia. The relevance of the treatment and management of wastewaters has augmented as a consequence of the increment in the number of biomedical residues that are thrown in cloacal stages. Frequently, these residues contain pathogens alive that could reach the water bodies, posing risks to human and environmental health (Iyer et al., 2021).
In the San Justo City, the first positive case for SARS-CoV-2, the etiological agent of the COVID-19 pandemic, was confirmed by reverse transcription-quantitative real-time PCR (RT-qPCR) on August 10, 2020 (https://www.santafe.gob.ar/ms/covid19/datosabiertos/).
It was reported that the SARS-CoV-2 can be found in the urine and feces of an infected person, even after the virus was not detected in the respiratory tract (Chen et al., 2020), and persist in the interior of fecal particles of domestic wastewater (DWW) (Arslan et al., 2020; Zhang et al., 2020). Another study showed that could remain viable, i.e., infectious, for up to 4.3 or 6 days in sewage or water, respectively (Giacobbo et al., 2021). It was communicated that over half of the 74 tested patients had stools that remained positive for SARS-CoV-2 RNA for an average of 11.2 days after the nasal swabs became negative (Wu et al., 2020).
This situation could be further aggravated in less developed countries, especially in areas with poor sanitation (Lodder and Roda Husman, 2020). Although important efforts have been made by the governments to limit the circulation of the virus, the transmission of SARS-CoV-2 may continue mainly due to the high infectiousness of new variants of the virus (Ullah et al., 2021). Recently, evidence has shown that contact with wastewater may lead to contamination (Giacobbo et al., 2021), but current knowledge does not confirm virus infectivity in wastewater yet, and knowledge gaps persist regarding its detection and the potential role of wastewater in its transmission (Kitajima et al., 2020).
RT-qPCR for diagnosis of the respiratory system through nasal swabs is an epidemiological tool developed and refined by environmental scientists over the last 20 years. WBE approach is now been used worldwide to understand the actual incidence of the COVID-19 pandemic, also standing out as an early-warning tool for predicting outbreaks in the future (Daughton, 2020; Sherchan et al., 2020; Chavarria-Miró et al., 2021; Xu et al., 2021a, 2021b; Ahmed et al., 2021, Randazzo et al., 2020a, 2020b; Lu et al., 2020; Orive et al., 2020; Bertels et al, 2022), besides many other ahead of print or preprints publications (Bloemen et al., 2022; Boehm et al., 2022; Hoar et al., 2022; Smith et al, 2022; Zhao et al., 2021; among others).
However, in South America, few studies have been reported (Fongaro et al. 2020; Michael-Kordatou et al., 2020; De Souza et al., 2020), and to the best of our knowledge, until now, five studies have been conducted to detect SARS-CoV-2 in Argentinian wastewaters (Iglesias et al. 2020; Barril et al. 2021; Giraud-Billoud et al., 2021; Barrios et al., 2021; Masachessi et al., 2022). Although the setup for the Argentinian treatment plants is different from the ones at San Justo City, none of them has studied the presence of SARS-CoV-2 RNA at different stages of DWW treatment.
In the present work, we sampled DWW at three stages of the treatment process: (1) untreated DWW; (2) treated DWW in secondary facultative lagoons; and (3) post-chlorinated DWW, to detect the presence/absence of viral RNA, to monitor physicochemical variables, and to check the effectiveness of the chlorination process.
We applied viral concentration methods and RNA isolation methods to each sample at different stages of the DWW treatment process to detect and quantity genetic markers of SARS-CoV-2 RNA by RT-qPCR and to confirm the effectiveness of chlorination as a pre-treatment step before sewage discharge into the river. In addition, we compared the temporal changes in the number of SARS-CoV-2 RNA genetic copies in DWW with the COVID-19 daily active cases reported in San Justo City. We expect that this approach could contribute to gaining knowledge on SARS-CoV-2 surveillance in DWW.
2 Materials and Methods
2.1 Study Area and Sample Collection
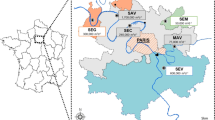
Santa Fe province is located in the center east of Argentina. In San Justo City, the city’s Public, Social, Social Assistance, and Housing Services Cooperative constructed the wastewater treatment plant (WWTP) and maintain the sewage network since 1998.
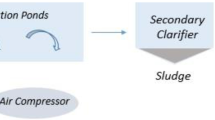
The samples were taken between July and August 2021 at the WWTP (30° 78′ 93″ S, 60° 62′ 09″ W). The WWTP processes a flow of 135 m3 h−1 of DWW from approximately 25,000 inhabitants. In addition, the WWTP receives the DWW brought by atmospheric trucks and from septic tanks located in areas of the city without a sewer network. The DWWs are treated by a stabilization and chlorination lagoon system before being discharged into the receiving aquatic environment, the Salado River (Fig. 1A). The treatment plant consists of (i) two anaerobic lagoons; (ii) two primary facultative lagoons; (iii) two secondary facultative lagoons; and (iv) a chlorination chamber.
To analyze SARS-CoV-2 RNA, ten 50-mL samples per sampling site (30 samples in total) at different sampling times between July and August 2021 were taken at three sampling sites, corresponding to the stages of the DWW treatment: site 1 (S1), untreated DWW, site 2 (S2), junction of two secondary facultative lagoons (prechlorination), and site 3 (S3), post-chlorination (Fig. 1B). Physicochemical and bacteriological parameters were also analyzed at each sampling site. No rainfall or storm events occurred on the sampling days or before them.
2.2 Physicochemical and Bacteriological Parameters of DWW
Some physicochemical and bacteriological parameters were recorded during the sampling period. At sites S1 and S2, the following parameters were determined: pH, chemical oxygen demand (COD, mg O2 L−1), biological oxygen demand (BOD, mg O2 L−1), sulfur (mg L−1), total suspended solids (TSS, mg L−1), nitrates (mg NO3− L−1), nitrites (mg NO2- L−1), ammonia (mg NH4+ L−1), phosphate (mg PO43− L−1), total coliform, and fecal bacteria (MPN 100 mL−1). At S3, all the mentioned parameters and residual chlorine (mg Cl2 L−1) were recorded. All analyses were made according to Standard Methods for the Examination of Water and Wastewater (APHA, 2017).
2.3 Pretreatment of Samples for SARS-CoV-2 RNA Detection
The DWW samples were stored in polypropylene containers and kept refrigerated (4 °C) until centrifugation and filtration (Róka et al. 2021; Kumar et al., 2020). The samples were centrifuged for 30 min at 4500 × g and filtered twice. For the first filtering, glass fiber filters (Sartorius MGC, 1.2-μm nominal pore size) and for the second filtering, Sartorius filters (0.2-μm nominal pore size) were used. Between filtering the samples, all the elements used were disinfected with a 10% sodium hypochlorite solution. After this process, all samples were stored at – 80 °C for 10 days until the analysis of SARS-CoV-2 RNA.
2.4 RNA Isolation and Extraction, SARS-CoV-2 Genetic Marker Detection and Quantification
RNA was extracted from the samples using a Thermo Scientific™ KingFisher automated system, with Invitrogen™ MagMAX™ kit for nucleic acid purification. Following Ahmed et al. (2020), sample volumes of 200 µL were extracted and incubated for 5 min at 65 °C with lysis buffer (265 µL of MagMAX™), proteinase K (5 µL), and magnetic beads (10 µL). After digestion, washes were performed with wash buffer 1 (500 µL) and 80% ethanol (500 µL) to obtain an RNA/DNA elution of 50 µL.
For the detection and quantification of viral RNA, PerkinElmer® SARS-CoV-2 real-time RT-PCR assay was used following the manufacturer’s instructions. The assay targets specific SARS-CoV-2 genomic regions: nucleocapsid gene (N) and open reading frame gene 1ab (ORF1ab). The assay is based on the TaqMan enzyme to perform in vitro viral RNA transcription, DNA amplification, and fluorescence detection. The two amplicons were labeled with FAM (N), HEX (ORF1ab), and internal control (Cy5) fluorescent dyes, respectively, to generate a target-specific signal.
The following mix was used for the PCR: CoV-2 reagent A-1 µL/test × n (samples); CoV-2 enzyme mix-5 µL/test × n (samples); and total-6 µL/test × n (samples). For a final volume of 20 µL, 14 µL of the nucleic acid extract to be studied was added to each mix. The COBAS z480 analyzer real-time thermal cycler (Roche®) was used. The cycling conditions were temperature (25 to 60 °C); time (2 min to 30 s); and 1 to 45 cycles. Nuclease-free water provided in the PerkinElmer® SARS-CoV-2 RT-qPCR reagent kit was used as a negative control. The negative control was added during extraction to each assay plate to control contamination during this step. Negative control PCR was performed using the same PCR mix used for the DWW samples. On the other hand, positive control was included in each extraction plate to ensure reagents and instruments perform optimally. The positive control was a plasmid containing target sequences of SARS-CoV-2: the ORF1ab and N gene of SARS-CoV-2. Both targets had Ct values of ≤ 35 and ≤ 32, respectively. The reaction mixture including a primer/probe set that detects a human RNA target was used as an internal control. The internal control is Cy5 fluorescent dye-labeled RNAse P (RPP30) and is used to monitor processes from nucleic acid extraction to fluorescence detection. RNAse P (internal control) should not be detected in either the positive control or the negative control. Positive and negative controls, performed with PerkinElmer® SARS-CoV-2 RT-qPCR reagent kit assay, are described in Table 1 (supplementary material).
The cycle threshold values (Ct) for two viral genes (N and ORF1ab) were considered positive if SARS-CoV-2 is detected when at least one target has Ct ≤ 40, following the instructions of the kit—PerkinElmer® SARS-CoV-2 real-time RT-PCR assay—and previous research (Giraud-Billoud et al. (2021) and Xu et al., (2021a, 2021b). The results were expressed as the number of genetic copies per 100 mL of DWW, and the conversion of Ct values to several copies per 100 mL−1 of the sample was carried out from emergency use authorization (USA) summary PerkinElmer SARS-CoV-2 RT-qPCR.
The evaluated linear dynamic range was between 60 and 3000 genetic copies of each marker per 100 mL. The standard curves had R2 = 0.891 (slope = − 309.97; y-intersect = 12.136) for the N marker and R2 = 0.763 (slope = − 406.01; y-intersect = 14.286) for ORF1ab marker. The results are expressed as the number of genetic copies per 100 mL DWW. Results of viral loads obtained from the dilutions performed for the genetic markers N and ORF1ab are shown in Table 2 (supplementary material).
2.5 Epidemiological Data
Data from daily clinically active cases were accessed through the COVID-19 Statistics Office of the local hospital, San Justo, Santa Fe. In addition, the number of accumulated cases was obtained from the Santa Fe Health Ministry (https://www.santafe.gob.ar/ms/covid19/datosabiertos/).
2.6 Data Analysis
To verify possible statistically significant differences between sampling sites (S1-S3) in the number of genetic copies of the two genetic markers used in this work, Kruskal–Wallis nonparametric tests were performed. Spearman’s correlation analyses were carried out to test correlations between COVID-19 daily clinically active cases and the number of genetic copies per 100 mL of DWW samples at the different sampling times and sampling sites (significance level at p < 0.05) by the GraphPad Prism 5.0 program (GraphPad Software Inc. n.d., San Diego, CA, USA).
3 Results
3.1 Physicochemical and Bacteriological Parameters in DWW
Table 1 shows the environmental parameters of each sampling site in the WWTP of San Justo City. All physicochemical parameters at S3 were within the mandatory limits for the disposal of sewage waste according to local regulations of the province of Santa Fe (ENRESS, 2011).
3.2 SARS-CoV-2 RNA Detection in DWW Samples
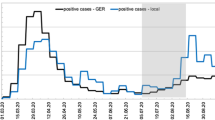
Out of the 30 analyzed samples, 30% were positive for SARS-CoV-2 RNA. From the total of positive samples (9 samples), 77% (7 samples) corresponded to S1 (untreated DWW), 23% (2 samples) to S2 (pre-chlorination), and no positivity for SARS-CoV-2 RNA was recorded at S3 (post-chlorination). Significant differences were observed in the number of positive samples between S1 and S2 (p < 0.05). The viral loads’ average number of genetic N per 100 mL−1 was as follows: at S1, 139,581 (min. = 92,659, max. = 179,850); at S2, 57,256 (min. = 4717, max. = 109,796), and at S3, the gen N dropped to 0. The ORF1ab gene viral load in S1 was average = 80,890, max. = 114,751, and min. = 31,925, but was not detected in S2 and S3 (Fig. 2, Table 2).
Results are reported for each of the two specific SARS-CoV-2 genomic regions: nucleocapsid gene (N) and open reading frame gene 1ab (ORF1ab). ––-, negative results.
On the other hand, in San Justo City, 179 clinical cases were reported during the period of the study. No correlation (p < 0.05) was found between the number of clinically active daily cases and the number of genetic copies of N per 100 mL−1 at S1, which is the sampling point with the highest positivity of cases.
In Fig. 2, it can be observed that the increase in the number of viral copies of the N gene could anticipate the number of clinical cases in the population. On August 7, an increase in clinical cases was detected (71 cases), and between 7 and 8 days before, on July 29 and 30, there was an increment in the number of SARS-CoV-2 RNA viral copies of the N gene 100 mL−1, augmenting from 129,474 N gene viral copies 100 mL−1 on July 28 to 179,850 and 175,200 N gene viral copies 100 mL−1 on July 29 and 30, respectively. At S2, the N gene was quantified only in 20% of the samples analyzed, and at S3 (post-chlorination), no viral RNA was detected.
4 Discussion
Since the emergence of the COVID-19 pandemic, many studies confirm that WBE is a useful tool that allows monitoring the progress of the pandemic and predicting increases in clinical cases, thus acting as an early alarm system for COVID-19 outbreak and as a complementary approach for public health monitoring at the population level (Tran et al. 2021; Gonçalves et al., 2021).
In our study, the DWW treatment under the stabilization lagoon system followed by chlorination (mean pH = 7.8 and 0.16 mg L−1 chlorine) was highly efficient in reducing and eliminating SARS-CoV-2 RNA, total coliforms from 2.40 × 105 to 200 MPN 100 mL−1, fecal coliforms from 9.30 × 104 to 40 MPN 100 mL−1, total coliforms diminished 45.8-folds and fecal coliforms 22.58-folds. In addition, we confirmed that the concentration of chlorine recommended by the WHO (≥ 0.5 mg L−1 after at least 30 min of contact time at pH 8.0) successfully removed SARS-CoV-2 RNA from DWW. In a similar work, the pathogenic microorganism concentrations in effluent achieved < 2 MPN L−1. The main disinfection was achieved at a chlorine dose of 2 mg L−1 (Zhang et al., 2020). The effect of chlorination was considered by several studies, typically in the context of biosafety of WWTP effluents (Bertels et al., 2022; Wigginton & Kohn, 2012; Wigginton et al., 2012). Regarding the mechanism involved, it has been reported that chlorine inactivates viruses through the cleavage of the capsid protein backbone of viruses, therefore inhibiting viral genome injection into host cells (Page et al., 2010). In another study, 19% out of 100 untreated wastewater samples taken from the primary influent contained SARS-CoV-2 RNA, while secondary and tertiary effluent samples were negative for SARS-CoV-2 RNA, indicating that the secondary and tertiary treatment processes were successful in removing SARS-CoV-2 RNA (Sherchan et al. 2020), as was also found in our survey.
Chlorine inactivation efficiency can be reduced because potential alternative hosts can store SARS-CoV-2 and function as reservoirs in wastewaters. It was reported that the SARS-CoV-2 can persist in the interior of fecal particles of domestic wastewater (DWW) (Arslan et al., 2020; Zhang et al., 2020). Alternative reservoirs of SARS-CoV-2 in WW are organic and suspended solids that can protect viruses that adsorb to these particles (Gundy et al., 2008). Conversely, many factors can modify the integrity of viruses in WW, e.g., pH temperature, solid phase/liquid phase, population number, dilution by rain, presence, and characteristics of organic matter and humic acids and the influence of microorganisms and biofilms, among other factors (Amoah et al., 2020; Bertels et al., 2022).
Some authors alert about the relevance of fecal shedding factors among other sewer system-dependent factors that are not fully understood (Bertels et al., 2022). In another study, Fores et al. (2021) observed that on average, 23% of SARS-CoV-2 N1 genes remain in the solid phase of wastewater after ultrafiltration. In this regard, Zhao et al. (2021) found that the supernatant fraction had high PCR threshold cycle (Ct) values, and most data were unquantifiable owing to low concentrations of SARS-CoV-2 RNA in the wastewater. In this work, we do not consider the fecal shedding factor. Future research should include SARS-CoV-2 loads in the solid fraction of raw influent, not only of the supernatant.
The minimum number of positive cases that there must be in a community for the virus to be detected in DWW is not yet exactly known, and a minimum number of infected people should be registered when trying to find correlations between concentrations of viral RNA and positive cases within a given population (Hong et al., 2021). The lack of correlation between the number of clinically active cases and the number of viral RNA copies of the gen N/100 mL−1 could be due to changes in the temperature of the DWW (Hart & Halden, 2020), the hydraulic retention time in the system, sampling times, chemical or biological compounds that may occur at specific times (Amoah et al., 2020), the extension of the sanitation network, variations in virus excretion (van Doorn et al., 2020), and the degradation of genetic material or the number of suspended solids (Mandal et al., 2020), or simply because the number of diagnosed cases was not high enough.
Hoar et al. (2022) recently published the results of an ongoing wastewater monitoring program that tracked trends in sewershed-level SARS-CoV-2 loads. An estimated minimum detectable case rate between 2 and 8 cases/day/100,000 people was associated with the method limit of detection in wastewater. In this sense, WBE is an attractive option for many municipalities, to confirm findings from clinical testing about population-level COVID-19 dynamics and to monitor for new outbreaks in instances when testing is inadequate.
Regarding the genetic markers selected to detect SARS-CoV-2-RNA in this work, -N1, N3,and ORB1ab genes-, these are also the most commonly found in the literature. The virus nucleocapsid (N) gene is widely used for WBE (Ahmed et al., 2021; Giraud-Billoud et al., 2021; Medema et al., 2020; Wu et al., 2020). In the detection of the SARS-CoV-2 RNA N gene, the Ct values found in this work were between 34.14 and 35.71. Similar Ct results for the N gene were reported by Medema et al. (2020), Pérez-Cataluña et al. (2021), and Giraud-Billoud et al. (2021) for DWW samples.
In general, OFR1ab genes were not as widely used as N genes to detect SARS-CoV-2 RNA. The results of our study show that the OFR1ab gene could be considered as a marker in WBE for COVID-19, but to confirm the use of this marker in WBE for COVID-19, more studies are needed. In another work, WBE surveillance through genetic material of SARS-CoV-2 viruses was also successfully applied. ORF1ab, N, and S genes of SARS-CoV-2 were found in the influent, with no genes being detected in the effluent (Kumar et al., 2020).
Considering the effectiveness of SARS-CoV-2 RNA quantification using RT-qPCR to anticipate the course of the pandemic, in our work, the rise in the SARS-CoV-2 genetic marker N occurred 6 days earlier than the rise in clinically active COVID-19 cases, even though more research has to be done to corroborate this result.
Similar findings revealed that the genetic markers N1 and N2 of SARS-CoV-2 increased between 3 and 6 days earlier than the confirmed cases of COVID-19 (Giraud-Billoud et al., 2021). In another study, the increase in SARS-CoV-2 RNA concentrations in primary sewage sludge was observed between 1 to 4 days before the admission of patients to local hospitals and 6 to 8 days before positive test results for SARS-CoV-2 (Peccia et al., 2020). Other studies reported that wastewater surveillance anticipated, by several weeks, not only the original COVID-19 pandemic wave but also the onset of the second wave (Chavarria-Miró et al., 2021), while Róka et al. (2021) detected the first positive signal in sewage 2 weeks before the rise in case of numbers could predict the second wave of the outbreak.
Another aspect to be considered is the observed variability in virus counting reported in the literature. In this line, a source of unexplained variability in SARS-CoV-2 surveys in WWTPs is presented by Bloemen et al. (2022). Disconnected sewer systems (e.g., septic tanks) reduce the number of persons connected to the sewer network. This is especially important for WWTPs in rural areas. In San Justo City, this variability was minimized because the WWTP receives the DWW brought by atmospheric trucks and from septic tanks located in areas of the city without a sewer network. No rainfall events occurred during the sampling period, thus excluding another source of potential variability.
The presence of SARS-CoV-2 virion-free RNA in urban wastewater from the city of San Justo was determined from two specific molecular markers. However, these results do not demonstrate the transmission capacity of SARS-CoV-2 from urban wastewater. Different authors have discussed the possibility of SARS-CoV-2 transmission to humans through sewage or through contact with surface water that receives untreated or inefficiently treated sewage. Ahmed et al., 2021, among many others). In this sense, Anand et al. (2022) reported that, depending on the fate and decomposition in wastewater, SARS-CoV-2 has a low risk of transmission to humans, but his review paper also shows that the knowledge of SARS-CoV-2 transmission routes and environmental fates still needs to be improved and that the effective surveillance and deactivation are critical. Although virion-free RNA was reported in this study, its deactivation allows us to hypothesize that it would not pose a risk to the environmental and human health of plant operators and users of the receiving water body (Salado River).
In synthesis, since the outbreak of the COVID pandemia, literature has accumulated considering WBE as a real-time low costing, noninvasive, and individual-independent methodology to track and prevent, to some extent, coronavirus events (Bertels et al., 2022). Several countries have successfully implemented WBE as a complementary system to clinical testing and acquired relevant qualitative and (semi-)quantitative data on community infection levels (Hasan et al., 2021).
Recent research considers that wastewater surveillance has become essential for monitoring the spread of SARS-CoV-2 (Pipes et al., 2022). Moreover, Smith et al. (2022) emphasize that publicly available health department incidence rates vastly underestimate true community incidence. Considering the relevance of rapid detection of the continuous appearance of new variants of the virus, it was documented that regional replacement of BA.1 with BA.2 is in agreement with, and ahead of, clinical sequencing data (Boehm et al., 2022).
Considering that wastewater represents a biological composite sample from the contributing community, continued population-level monitoring from wastewater data could become increasingly useful in areas where clinical testing rates decline or resources for clinical testing are limited (Hoar et al., 2022). In this line, this work could contribute to building a network for WBE to monitor SARS-CoV-2 RNA in wastewaters during the pandemic waves and the post-pandemic.
5 Conclusions
-
- The presence of SARS-CoV-2 virion-free RNA in urban wastewater was determined from two specific molecular markers.
-
- The viral loads of N and OFR1ab genes decreased significantly between S1 and S2, and the number of positive samples decreased as follows: S1>S2>S3, finally eliminating SARS-CoV-2 RNA.
-
- Results presented here can assist local authorities in taking action on public health, enabling the cooperation between research and management institutions.
-
- Assessment of wastewater matrix effects should be considered for future research aiming to characterize uncertainty in WBE data. It is recommended that solid-phase analyses should complement aqueous phase samples.
-
- The results of this work contribute to the WBE for COVID-19, confirming that it is an early warning system, which allows detecting if the virus is in a community, based on its presence, concentrations, or absence, in the different stages of the DWW treatment.
Data Availability
Data on this paper can be found at https://docs.google.com/spreadsheets/d/1tl3FBurgYd-T-0bYyDgebQ_BydAJBZjc/edit?usp=sharing&ouid=102665681467869564680&rtpof=true&sd=true.
References
Ahmed, W., Bivins, A., Bertsch, P. M., Bibby, K., Choi, P. M., Farkas, K., Gyawali, P., Hamilton, K. A., Haramoto, E., Kitajima, M., Simpson, S. L., Tandukar, S., Thomas, K., & Mueller, J. F. (2020). Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimization and quality control are crucial for generating reliable public health information. Current Opinion in Environmental Science & Health. https://doi.org/10.1016/j.coesh.2020.09.003
Ahmed, F., Islam, M. A., Kumar, M., Hossain, M., Bhattacharya, P., Islam, M. T., Hossen, F., Hossain, M. S., Islam, M. S., Uddin, M. M., Islam, M. N., Bahadur, N. M., Didar-ul-Alam, M., Reza, H. M., & Jakariya, M. (2021). First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation centre in Bangladesh: Variation along the sewer network. Science of the Total Environment, 776, 145724. https://doi.org/10.1016/j.scitotenv.2021.145724
American Public Health Association. (2017). Standard Methods for the Examination of Water and Wastewater. In L. Baird, R. B., Eaton, A. D., Rice, E. W., & Bridgewater (eds.), (23rd ed.).
Amoah, I. D., Kumari, S., & Bux, F. (2020). Coronaviruses in wastewater processes: Source, fate, and potential risks. Environment International, 143(July), 105962. https://doi.org/10.1016/j.envint.2020.105962
Anand, U., Li, X., Sunita, K., Lokhandwala, S., Gautam, P., Suresh, S., et al. (2022). SARS-CoV-2 and other pathogens in municipal wastewater, landfill leachate, and solid waste: A review about virus surveillance, infectivity, and inactivation. Environmental Research, 203(July 2021), 111839. https://doi.org/10.1016/j.envres.2021.111839
Arslan, M., Xu, B., & Gamal El-Din, M. (2020). Transmission of SARS-CoV-2 via fecal-oral and aerosols–borne routes: Environmental dynamics and implications for wastewater management in underprivileged societies. Science of the Total Environment, 743, 140709. https://doi.org/10.1016/j.scitotenv.2020.140709
Barril, P. A., Pianciola, L. A., Mazzeo, M., Ousset, M. J., Jaureguiberry, M. V., Alessandrello, M., Sánchez, G., & Oteiza, J. M. (2021). Evaluation of viral concentration methods for SARS-CoV-2 recovery from wastewaters. Science of the Total Environment, 756, 144105. https://doi.org/10.1016/j.scitotenv.2020.144105
Barrios, M. E., Díaz, S. M., Torres, C., Costamagna, D. M., Blanco Fernández, M. D., & Mbayed, V. A. (2021). Dynamics of SARS-CoV-2 in wastewater in three districts of the Buenos Aires metropolitan region, Argentina, throughout nine months of surveillance: A pilot study. Science of the Total Environment, 800, 149578. https://doi.org/10.1016/j.scitotenv.2021.149578
Bertels, X., Demeyer, P., Van den Bogaert, S., Boogaerts, T., van Nuijs, A. L. N., & Delputte, P. (2022). Lahousse L (2022) Factors influencing SARS-CoV-2 RNA concentrations in wastewater up to the sampling stage: A systematic review. Science of the Total Environment, 820, 153290.
Bloemen, M., Delang, L., Rector, A., Raymenants, J., Thibaut, J., Pussig, B., Fondu, L., Aertgeerts, B., Van Ranst, M., Van Geet,C., Arnout, J., & Wollants E. (2022). Detection Of SARS-COV-2 Variants Of Concern In Wastewater Of Leuven, Belgium. medRxiv 2022.05.12.22274823; https://doi.org/10.1101/2022.05.12.22274823
Boehm, AB, Hughes, B., Wolfe, M.K., White, B J., Duong, D., & Chan-Herur, V. (2022). Regional replacement of SARS-CoV-2 variant BA.1 with BA.2 as observed through wastewater surveillance. medRxiv 2022.04.22.22274160; https://doi.org/10.1101/2022.04.22.22274160
Chavarria-Miró, G., Anfruns-Estrada, E., Martínez-Velázquez, A., Vázquez-Portero, M., Guix, S., Paraira, M., Galofré, B., Sánchez, G., Pintó, R. M., & Bosch, A. (2021). Time evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in wastewater during the first pandemic wave of COVID-19 in the metropolitan area of Barcelona Spain. Applied and Environmental Microbiology, 87(7), 1–9. https://doi.org/10.1128/AEM.02750-20
Chen, Chen, Gao, G., Xu, Y., Pu, L., Wang, Q., & Wang, L. (2020). SARS-CoV-2–positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Annals of Internal Medicine, 172(1), ITC1–ITC14. https://doi.org/10.7326/AWED202001070
Daughton, C. G. (2020). Wastewater surveillance for population-wide Covid-19: The present and future. Science of the Total Environment, 736, 139631. https://doi.org/10.1016/j.scitotenv.2020.139631
De Souza, C. D. F., Machado, M. F., & Do Carmo, R. F. (2020). Human development, social vulnerability and COVID-19 in Brazil: A study of the social determinants of health. Infectious Diseases of Poverty, 9(1), 4–13. https://doi.org/10.1186/s40249-020-00743-x
ENRESS (2011). Quality control regulation of sewage eflueents and disposal of biosolids, 8 (2011). http://www.enress.gov.ar/prestadores/otros_prestadores/resoluciones_regulatorias/docs/resoluciones/resoluciones_regulatorias/2011/324.pdf
Fongaro, G., Stoco, P. H., Souza, D. S. M., Grisard, E. C., Magri, M. E., Rogovski, P., Schörner, M. A., Barazzetti, F. H., Christoff, A. P., de Oliveira, L. F. V., Bazzo, M. L., Wagner, G., Hernández, M., & Rodriguez-Lázaro, D. (2020). SARS-CoV-2 in human sewage in Santa Catalina, Brazil, November 2019. MedRxiv, November 2019. https://doi.org/10.1101/2020.06.26.20140731
Fores, E., Bofill-Mas, S., Itarte, M., Martinez-Puchol, S., Hundesa, A., Calvo, M., Borrego, C. M., Corominas, L. L., Girones, R., & Rusinol, M. (2021). Evaluation of two rapid ultrafiltration based methods for SARS-CoV-2 concentration from wastewater. Science of the Total Environment, 768, 144786. https://doi.org/10.1016/j.scitotenv.2020.144786
Giacobbo, A., Rodrigues, M. A. S., Zoppas Ferreira, J., Bernardes, A. M., & de Pinho, M. N. (2021). A critical review on SARS-CoV-2 infectivity in water and wastewater. What do we know? Science of the Total Environment, 774, 145721. https://doi.org/10.1016/j.scitotenv.2021.145721
Giraud-Billoud, M., Cuervo, P., Altamirano, J. C., Pizarro, M., Aranibar, J. N., Catapano, A., Cuello, H., Masachessi, G., & Vega, I. A. (2021). Monitoring of SARS-CoV-2 RNA in wastewater as an epidemiological surveillance tool in Mendoza Argentina. Science of the Total Environment, 796, 148887. https://doi.org/10.1016/j.scitotenv.2021.148887
Gonçalves, J., Koritnik, T., Mioč, V., Trkov, M., Bolješič, M., Berginc, N., Prosenc, K., Kotar, T., & Paragi, M. (2021). Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Science of the Total Environment, 755, 4–10. https://doi.org/10.1016/j.scitotenv.2020.143226
GraphPad Software, Inc. (San Diego, CA, USA) version 7.0
Gundy, P. M., Gerba, C. P., & Pepper, I. L. (2008). Survival of coronaviruses in water and wastewater. Food and Environmental Virology, 1(1), 10.
Hart, O. E., & Halden, R. U. (2020). Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities, and challenges. Science of the Total Environment, 730, 138875. https://doi.org/10.1016/j.scitotenv.2020.138875
Hasan, S. W., Ibrahim, Y., Daou, M., Kannout, H., Jan, N., Lopes, A., Alsafar, H., & Yousef, A. F. (2021). Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: Surveillance of COVID-19 epidemic in the United Arab Emirates. Science of the Total Environment, 764, 142929. https://doi.org/10.1016/j.scitotenv.2020.142929
Hoar, C., Chauvin, F., Clare, A., McGibbon, H., Castro, E., Patinella, S., Katehis, D., Dennehy, J.J., Trujillo, M., Smyth, D S., & Silverman A.I. (2022). Monitoring SARS-CoV-2 in wastewater during New York City’s second wave of COVID-19: Sewershed-level trends and relationships to publicly available clinical testing data. medRxiv 2022.02.08.22270666; https://doi.org/10.1101/2022.02.08.22270666
Hong, P. Y., Rachmadi, A. T., Mantilla-Calderon, D., Alkahtani, M., Bashawri, Y. M., Al Qarni, H., O’Reilly, K. M., & Zhou, J. (2021). Estimating the minimum number of SARS-CoV-2 infected cases needed to detect viral RNA in wastewater: To what extent of the outbreak can surveillance of wastewater tell us? Environmental Research, 195, 110748. https://doi.org/10.1016/j.envres.2021.110748
Iglesias, N. G., Gebhard, L. G., Carballeda, J. M., Aiello, I., Recalde, E., Terny, G., Ambrosolio, S., L’Arco, G., Konfino, J., & Brardinelli, J. I. (2020). SARS-CoV-2 surveillance in untreated wastewater: First detection in a low-resource community in Buenos Aires, Argentina. MedRxiv, 1–7. https://doi.org/10.1101/2020.10.21.20215434
Iyer, M., Tiwari, S., Renu, K., Pasha, M. Y., Pandit, S., Singh, B., et al. (2021). Environmental survival of SARS-CoV-2 – A solid waste perspective. Environmental Research, 197(March), 111015. https://doi.org/10.1016/j.envres.2021.111015
Kitajima, M., Ahmed, W., Bibby, K., Carducci, A., Gerba, C. P., Hamilton, K. A., Haramoto, E., & Rose, J. B. (2020). SARS-CoV-2 in wastewater: State of the knowledge and research needs. Science of the Total Environment, 739, 139076. https://doi.org/10.1016/j.scitotenv.2020.139076
Kumar, M., Patel, A. K., Shah, A. V., Raval, J., Rajpara, N., Joshi, M., & Joshi, C. G. (2020). First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Science of the Total Environment, 746, 141326. https://doi.org/10.1016/j.scitotenv.2020.141326
Lodder, W., & de Roda Husman, A. M. (2020). SARS-CoV-2 in wastewater: Potential health risk, but also data source. The Lancet Gastroenterology and Hepatology, 5(6), 533–534. https://doi.org/10.1016/S2468-1253(20)30087-X
Lu, D., Huang, Z., Luo, J., Zhang, X., & Sha, S. (2020). Primary concentration – The critical step in implementing the wastewater-based epidemiology for the COVID-19 pandemic: A mini-review. Science of the Total Environment, 747, 141245. https://doi.org/10.1016/j.scitotenv.2020.141245
Mandal, P., Gupta, A. K., & Dubey, B. K. (2020). A review on presence, survival, disinfection/removal methods of coronavirus in wastewater and progress of wastewater-based epidemiology. Journal of Environmental Chemical Engineering, 8(5), 104317. https://doi.org/10.1016/j.jece.2020.104317
Masachessi, G., Castro, G., Cachi, A. M., Marinzalda, M. A., Liendo, M., Pisano, M. B., Sicilia, P., Ibarra, G., Rojas, R. M., Lopez, L., Barbas, G., Cardozo, D., Re, V. E., & Nates, S. V. (2022). Wastewater-based epidemiology as a silent sentinel of the trend of SARS-CoV-2 circulation in the community in central Argentina. Water Research, 219, 118541.
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., & Brouwer, A. (2020). Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environmental Science and Technology Letters, 7(7), 511–516. https://doi.org/10.1021/acs.estlett.0c00357
Michael-Kordatou, I., Karaolia, P., & Fatta-Kassinos, D. (2020). Sewage analysis as a tool for the COVID-19 pandemic response and management: The urgent need for optimized protocols for SARS-CoV-2 detection and quantification. Journal of Environmental Chemical Engineering, 8(5), 104306. https://doi.org/10.1016/j.jece.2020.104306
Núñez-Delgado, A., Zhou, Y., & Domingo, J. L. (2021). Editorial of the VSI “Environmental, ecological and public health considerations regarding coronaviruses, other viruses, and other microorganisms potentially causing pandemic diseases.” Environmental Research, 192, 110322. https://doi.org/10.1016/j.envres.2020.110322
Orive, G., Lertxundi, U., & Barcelo, D. (2020). Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Science of the Total Environment., 732, 139298. https://doi.org/10.1016/j.scitotenv.2020.139298
Page, M. A., Shisler, J. L., & Marin, B. J. (2010). Mechanistic aspects of adenovirus serotype 2 inactivation with free chlorine. Applied and Environmental Microbiology, 76, 2946–2954. https://doi.org/10.1128/AEM.02267-09 American Society for Microbiology.
Peccia, J., Zulli, A., Brackney, D. E., Grubaugh, N. D., Kaplan, E. H., Casanovas-Massana, A., Ko, A. I., Malik, A. A., Wang, D., Wang, M., Warren, J. L., Weinberger, D. M., Arnold, W., & Omer, S. B. (2020). Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nature Biotechnology, 38(10), 1164–1167. https://doi.org/10.1038/s41587-020-0684-z
Pérez-Cataluña, A., Cuevas-Ferrando, E., Randazzo, W., Falcó, I., Allende, A., & Sánchez, G. (2021). Comparing analytical methods to detect SARS-CoV-2 in wastewater. Science of the Total Environment, 758, 143870. https://doi.org/10.1016/j.scitotenv.2020.143870
Pipes L, Chen Z, Afanaseva S, Nielsen R. (2022) Estimating the relative proportions of SARS-CoV-2 strains from wastewater samples. medRxiv 2022.01.13.22269236; https://doi.org/10.1101/2022.01.13.22269236
Randazzo, W., Cuevas-Ferrando, E., Sanjuán, R., Domingo-Calap, P., & Sánchez, G. (2020a). 713 Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. International Journal of Hygiene and Environmental Health, 230, 1–8. https://doi.org/10.1016/j.ijheh.2020.113621715
Randazzo, W., Cuevas-Ferrando, E., Sanjuán, R., Domingo-Calap, P., & Sánchez, G. (2020). Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. International Journal of Hygiene and Environmental Health, 230, 113621. https://doi.org/10.1016/j.ijheh.2020b.113621
Róka, E., Khayer, B., Kis, Z., Kovács, L. B., Schuler, E., Magyar, N., Málnási, T., Oravecz, O., Pályi, B., Pándics, T., & Vargha, M. (2021). Ahead of the second wave: Early warning for COVID-19 by wastewater surveillance in Hungary. Science of the Total Environment, 786, 147398. https://doi.org/10.1016/j.scitotenv.2021.147398
Sherchan, S. P., Shahin, S., Ward, L. M., Tandukar, S., Aw, T. G., Schmitz, B., Ahmed, W., & Kitajima, M. (2020). First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Science of the Total Environment, 743, 140621. https://doi.org/10.1016/j.scitotenv.2020.140621
Smith, T., Holm, R.H., Keith, R.J., Amraotkar, A.R., Alvarado, C.R., Banecki, K., Choi, B, Santisteban, I. C., Bushau-Sprinkle, A.M., Kitterman, K.T., Fuqua, J., Hamorsky, K.T., Palmer, K.E., Brick, J.M., Rempala, G.A., & Bhatnagar A. (2022). Quantifying the relationship between sub-population wastewater samples and community-wide SARS-CoV-2 seroprevalence medRxiv 2022.04.28.22274086; https://doi.org/10.1101/2022.04.28.22274086
Tran, H. N., Le, G. T., Nguyen, D. T., Juang, R. S., Rinklebe, J., Bhatnagar, A., Lima, E. C., Iqbal, H. M. N., Sarmah, A. K., & Chao, H. P. (2021). SARS-CoV-2 coronavirus in water and wastewater: A critical review about presence and concern. Environmental Research, 193(September 2020). https://doi.org/10.1016/j.envres.2020.110265
Ullah, A., Mabood, N., Maqbool, M., Khan, L., Khan, M., & Ullah, M. (2021). SAR-CoV-2 infection, emerging new variants, and the role of activation-induced cytidine deaminase (AID) in lasting immunity. Saudi Pharmaceutical Journal, 29(10), 1181–1184. https://doi.org/10.1016/j.jsps.2021.09.006
van Doorn, A. S., Meijer, B., Frampton, C. M. A., Barclay, M. L., & de Boer, N. K. H. (2020). Systematic review with meta-analysis: SARS-CoV-2 stool testing and the potential for faecal-oral transmission. Alimentary Pharmacology and Therapeutics, 52(8), 1276–1288. https://doi.org/10.1111/apt.16036
Wigginton, K. R., & Kohn, T. (2012). Virus disinfection mechanisms: The role of virus composition, structure, and function. Current Opinion in Virology, 2(1), 84–89. https://doi.org/10.1016/j.coviro.2011.11.003
Wigginton, K. R., Pecson, B. M., Sigstam, T., Bosshard, F., & Kohn, T. (2012). Virus inactivation mechanisms: Impact of disinfectants on virus function and structural integrity. Environmental Science & Technology, 46(21), 12069–12078. https://doi.org/10.1021/es3029473
Wu, Y., Guo, C., Tang, L., Hong, Z., Zhou, J., Dong, X., Yin, H., Xiao, Q., Tang, Y., Qu, X., Kuang, L., Fang, X., Mishra, N., Lu, J., Shan, H., Jiang, G., & Huang, X. (2020). Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. The Lancet Gastroenterology and Hepatology, 5(5), 434–435. https://doi.org/10.1016/S2468-1253(20)30083-2
Xu, X., Zheng, X., Li, S., Lam, N. S., Wang, Y., Chu, D. K. W., Poon, L. L. M., Tun, H. M., Peiris, M., Deng, Y., Leung, G. M., & Zhang, T. (2021). The first case study of wastewater-based epidemiology of COVID-19 in Hong Kong. Science of the Total Environment, 790, 148000. https://doi.org/10.1016/j.scitotenv.2021.148000
Xu, X., Zheng, X., Li, S., Lam, N. S., Wang, Y., Chu, D. K. W., Poon, L. L. M., Tun, H. M., Peiris, M., Deng, Y., Leung, G. M., & Zhang, T. (2021). The first case study of wastewater-based epidemiology of COVID-19 in Hong Kong. Science of the Total Environment, 790, 148000. https://doi.org/10.1016/j.scitotenv.2021.148000
Zhang, D., Ling, H., Huang, X., Li, J., Li, W., Yi, C., Zhang, T., Jiang, Y., He, Y., Deng, S., Zhang, X., Wang, X., Liu, Y., Li, G., & Qu, J. (2020). Potential spreading risks and disinfection challenges of medical wastewater by the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Science of the Total Environment, 741, 140445. https://doi.org/10.1016/j.scitotenv.2020.140445
Zhao, B., Yu, Z., Fujita, T., Nihei, Y., Tanaka, H., & Ihara M.(2021) Tracking community infection dynamics of COVID-19 by monitoring SARS-CoV-2 RNA in wastewater, counting positive reactions by qPCR medRxiv 2021.12.23.21268343; https://doi.org/10.1101/2021.12.23.21268343
Acknowledgements
Special thanks go to the Public, Social, Social Assistance, and Housing Services Cooperative of San Justo, to the Government and Hospital “Dr. Juan O. Lanza” of the San Justo City, and the Central Laboratory of the Ministry of Health, Santa Fe, Argentina. The Central Laboratory is part of the Provincial Network of Laboratories of the Santa Fe Province and provides services of different complexity and specificity: biochemical, pathological anatomy, chemotherapy, immunology, genetics, microbiology, and monitoring of the COVID-19 pandemic. The laboratory is equipped with a Class II Biological Safety Cabinet (Code ECSB003) and has a national certification number N° C0521822-01.
Funding
The work was supported by the “Agencia Nacional de Promoción Científica y Tecnológica” PICT 2019 N° 02591. The project director is the researcher Dr. Luciana Regaldo.
Author information
Authors and Affiliations
Contributions
Ulises Reno: Conceptualization, Methodology, Formal analysis, Investigation, Writing-original draft preparation, Writing-review and editing. Luciana Regaldo: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Writing-review and editing. Guillermo Ojeda: Methodology, Validation, Formal analysis, Investigation. Josefina Schmuck: Writing-review and editing, Visualization. Natalí Romero: Writing-review and editing, Visualization. Wanda Polla: Writing-review and editing. Vanesa Kergaravat: Writing-review and editing. Ana María Gagneten: Conceptualization, Investigation, Original draft preparation, Writing-review and editing, Supervision.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Reno, U., Regaldo, L., Ojeda, G. et al. Wastewater-Based Epidemiology: Detection of SARS-CoV-2 RNA in Different Stages of Domestic Wastewater Treatment in Santa Fe, Argentina. Water Air Soil Pollut 233, 372 (2022). https://doi.org/10.1007/s11270-022-05772-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-022-05772-w