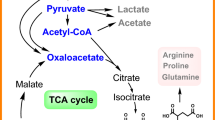
Abstract
Bacillus subtilis is a natural producer of 2,3-butanediol (2,3-BDO) and has acquired “Generally Regarded as Safe" status. It is reported to produce 2,3-BDO from synthetic sugars as well as complex and economic sugar sources such as molasses. However, the rate-limiting step in the formation of 2,3-BDO is its conversion from acetoin to 2,3-BDO by the enzyme butanediol dehydrogenase (2,3-BDH). Such 2,3-BDHs were screened based on higher affinity (lower Km) towards acetoin as substrate. The in silico docking studies were conducted for further validation, and they showed a high interaction profile for the PpBDH protein towards acetoin. Heterologous expression of these genes was studied in engineered Bacillus subtilis (BS1A1). In this study, it was seen that 2,3-BDH from Paenibacillus polymyxa ZJ-9 was reported to have higher enzyme activity levels, and in the fermentation studies, it was seen that the ratio of 2,3-BDO to acetoin was increased by 80.25%. The insights encourage further bioprocess optimization for increasing the fermentative production of 2,3-BDO. Our results provide a potential strategy to avoid the back conversion of 2,3-BDO to acetoin in an engineered Bacillus system.

















Similar content being viewed by others
Data availability
All the research data generated from this study and the biological data used from NCBI has been mentioned and cited with references in the manuscript.
References
Data Bridge Market Research (2020) Renewable Chemicals Market – Global Industry Trends and Forecast to 2029. https://www.databridgemarketresearch.com/reports/global-renewable-chemicals-market. Accessed: 10 Mar 2022
Celińska E, Grajek W (2009) Biotechnological production of 2,3-butanediol–current state and prospects. Biotechnol Adv 27:715–725. https://doi.org/10.1016/j.biotechadv.2009.05.002
Białkowska AM (2016) Strategies for efficient and economical 2,3-butanediol production: new trends in this field. World J Microbiol Biotechnol 32:200. https://doi.org/10.1007/s11274-016-2161-x
Qi G, Kang Y, Li L et al (2014) Deletion of meso-2,3-butanediol dehydrogenase gene budC for enhanced D-2,3-butanediol production in Bacillus licheniformis. Biotechnol Biofuels 7:16. https://doi.org/10.1186/1754-6834-7-16
Białkowska AM, Gromek E, Krysiak J et al (2015) Application of enzymatic apple pomace hydrolysate to production of 2,3-butanediol by alkaliphilic Bacillus licheniformis NCIMB 8059. J Ind Microbiol Biotechnol 42:1609–1621. https://doi.org/10.1007/s10295-015-1697-3
Keo-Oudone C, Phommachan K, Suliya O et al (2022) Highly efficient production of 2,3-butanediol from xylose and glucose by newly isolated thermotolerant Cronobacter sakazakii. BMC Microbiol 22:164. https://doi.org/10.1186/s12866-022-02577-z
Hazeena SH, Sindhu R, Pandey A et al (2020) Lignocellulosic bio-refinery approach for microbial 2,3-Butanediol production. Bioresour Technol 302:122873. https://doi.org/10.1016/j.biortech.2020.122873
Soltys KA, Batta AK, Koneru B (2001) Successful nonfreezing, subzero preservation of rat liver with 2,3-butanediol and type I antifreeze protein. J Surg Res 96:30–34. https://doi.org/10.1006/jsre.2000.6053
Gräfje H, Körnig W, Weitz HM et al (2000) Butanediols, butenediol, and butynediol. Ullmann’s Encycl Ind Chem. https://doi.org/10.1002/14356007.a04_455
Transparency Market Research (2021) 2,3-Butanediol Market. https://www.transparencymarketresearch.com/2-3-butanediol-market.html. Accessed 3 May 2021
Ma C, Wang A, Qin J et al (2009) Enhanced 2,3-butanediol production by Klebsiella pneumoniae SDM. Appl Microbiol Biotechnol 82:49–57. https://doi.org/10.1007/s00253-008-1732-7
Ji XJ, Huang H, Zhu JG et al (2010) Engineering Klebsiella oxytoca for efficient 2, 3-butanediol production through insertional inactivation of acetaldehyde dehydrogenase gene. Appl Microbiol Biotechnol 85:1751–1758. https://doi.org/10.1007/s00253-009-2222-2
Huynh DT, Kim AY, Seol IH et al (2015) Inactivation of the virulence factors from 2,3-butanediol-producing Klebsiella pneumoniae. Appl Microbiol Biotechnol 99:9427–9438. https://doi.org/10.1007/s00253-015-6861-1
Indian Council of Agricultural Research Biosafety Portal (2023) Recombinant DNA Safety Guidelines, 1990. https://biosafety.icar.gov.in/recombinant-dna-safety-guidelines-1990-2/. Accessed 26 Oct 2023
Yang Z, Zhang Z (2019) Recent advances on production of 2, 3-butanediol using engineered microbes. Biotechnol Adv 37:569–578. https://doi.org/10.1016/j.biotechadv.2018.03.019
Kim SJ, Kim JW, Lee YG et al (2017) Metabolic engineering of Saccharomyces cerevisiae for 2,3-butanediol production. Appl Microbiol Biotechnol 101:2241–2250. https://doi.org/10.1007/s00253-017-8172-1
Cui W, Han L, Suo F et al (2018) Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J Microbiol Biotechnol 34:145. https://doi.org/10.1007/s11274-018-2531-7
Pohl S, Harwood CR (2010) Heterologous protein secretion by bacillus species from the cradle to the grave. Adv Appl Microbiol 73:1–25. https://doi.org/10.1016/S0065-2164(10)73001-X
Zhang J, Zhao X, Zhang J et al (2017) Effect of deletion of 2,3-butanediol dehydrogenase gene (bdhA) on acetoin production of Bacillus subtilis. Prep Biochem Biotechnol 47:761–767. https://doi.org/10.1080/10826068.2017.1320293
Wang D, Oh BR, Lee S et al (2021) Process optimization for mass production of 2,3-butanediol by Bacillus subtilis CS13. Biotechnol Biofuels 14:15. https://doi.org/10.1186/s13068-020-01859-w
Deshmukh AN, Mistry SR, Yewale TB et al (2015) Production of 2, 3-butanediol from sugarcane molasses using Bacillus subtilis. Int J Adv Biotechnol Res 6:66–79
Maina S, Prabhu AA, Vivek N et al (2022) Prospects on bio-based 2,3-butanediol and acetoin production: recent progress and advances. Biotechnol Adv 54:107783. https://doi.org/10.1016/j.biotechadv.2021.107783
Lee GB, Kim YJ, Lim JK et al (2019) A simple biosynthetic pathway for 2,3-butanediol production in Thermococcus onnurineus NA1. Appl Microbiol Biotechnol 103:3477–3485. https://doi.org/10.1007/s00253-019-09724-z
Nicholson WL (2008) The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl Environ Microbiol 74:6832–6838. https://doi.org/10.1128/AEM.00881-08
Zhang X, Zhang R, Yang T et al (2013) Mutation breeding of acetoin high producing Bacillus subtilis blocked in 2,3-butanediol dehydrogenase. World J Microbiol Biotechnol 29:1783–1789. https://doi.org/10.1007/s11274-013-1339-8
Hajieghrari B, Farrokhi N, Goliaei B et al (2015) Computational identification, characterization and analysis of conserved miRNAs and their targets in Amborella Trichopoda. J Data Mining Genomics Proteomics 6:168. https://doi.org/10.4172/2153-0602.1000168
Mirdita M, Schütze K, Moriwaki Y et al (2022) ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. https://doi.org/10.1038/s41592-022-01488-1
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. https://doi.org/10.1002/pro.5560020916
Dundas J, Ouyang Z, Tseng J et al (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34:W116–W118. https://doi.org/10.1093/nar/gkl282
Yende SR, Shah SK, Arora SK et al (2021) In silico prediction of phytoconstituents from Ehretia laevis targeting TNF-α in arthritis. Digit Chin Med 4:180–190. https://doi.org/10.1016/j.dcmed.2021.09.003
Abdelkader A, Elzemrany AA, El-Nadi M et al (2022) In-silico targeting of SARS-CoV-2 NSP6 for drug and natural products repurposing. Virology 573:96–110. https://doi.org/10.1016/j.virol.2022.06.008
Altuner EM (2022) In silico proof of the effect of quercetin and umbelliferone as alpha-amylase inhibitors, which can be used in the treatment of diabetes. Kastamonu Uni Orman Fakültesi Dergisi 22:202–216. https://doi.org/10.17475/kastorman.1215281
Khaerunnisa S, Kurniawan H, Awaluddin R et al (2020) Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints. https://doi.org/10.20944/preprints202003.0226.v1
Wickham H (2016) Data analysis. In: Wickham H (ed) ggplot2, 2nd edn. Springer, Cham
Guérout-Fleury AM, Frandsen N, Stragier P (1996) Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61. https://doi.org/10.1016/s0378-1119(96)00404-0
Vojcic L, Despotovic D, Martinez R et al (2012) An efficient transformation method for Bacillus subtilis DB104. Appl Microbiol Biotechnol 94:487–493. https://doi.org/10.1007/s00253-012-3987-2
Kaltwasser M, Wiegert T, Schumann W (2002) Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl Environ Microbiol 68:2624–2628. https://doi.org/10.1128/AEM.68.5.2624-2628.2002
Yang TW, Rao ZM, Zhang X et al (2013) Effects of corn steep liquor on production of 2,3-butanediol and acetoin by Bacillus subtilis. Process Biochem 48:1610–1617. https://doi.org/10.1016/j.procbio.2013.07.027
Deshmukh AN, Nipanikar-Gokhale P, Jain R (2016) Engineering of Bacillus subtilis for the production of 2,3-butanediol from sugarcane molasses. Appl Biochem Biotechnol 179:321–331. https://doi.org/10.1007/s12010-016-1996-9
Hamza TA, Hadwan MH (2020) New spectrophotometric method for the assessment of catalase enzyme activity in biological tissues. Curr Anal Chem 16:1054–1062. https://doi.org/10.2174/1573411016666200116091238
Yan Y, Lee CC, Liao JC (2009) Enantioselective synthesis of pure (R, R)-2,3-butanediol in Escherichia coli with stereospecific secondary alcohol dehydrogenases. Org Biomol Chem 7:3914–3917. https://doi.org/10.1039/b913501d
Muschallik L, Molinnus D, Bongaerts J et al (2017) (R, R)-Butane-2,3-diol dehydrogenase from Bacillus clausii DSM 8716T: cloning and expression of the bdhA-gene, and initial characterization of enzyme. J Biotechnol 258:41–50. https://doi.org/10.1016/j.jbiotec.2017.07.020
Gao J, Yang HH, Feng XH et al (2013) A 2,3-butanediol dehydrogenase from Paenibacillus polymyxa ZJ-9 for mainly producing R, R-2,3-butanediol: purification, characterization and cloning. J Basic Microbiol 53:733–741. https://doi.org/10.1002/jobm.201200152
Zhang GL, Wang CW, Li C (2012) Cloning, expression and characterization of meso-2,3-butanediol dehydrogenase from Klebsiella pneumoniae. Biotechnol Lett 34:1519–1523. https://doi.org/10.1007/s10529-012-0933-4
Xu GC, Bian YQ, Han RZ et al (2016) Cloning, expression, and characterization of budC gene encoding meso-2,3-butanediol dehydrogenase from Bacillus licheniformis. Appl Biochem Biotechnol 178:604–617. https://doi.org/10.1007/s12010-015-1897-3
Ui S, Okajima Y, Mimura A et al (1997) Sequence analysis of the gene for and characterization of D-acetoin forming meso-2,3-butanediol dehydrogenase of Klebsiella pneumoniae expressed in Escherichia coli. J Biosci Bioeng 83:32–37. https://doi.org/10.1016/S0922-338X(97)87323-0
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Auld DS, Bergman T (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: the role of zinc for alcohol dehydrogenase structure and function. Cell Mol Life Sci 65:3961–3970. https://doi.org/10.1007/s00018-008-8593-1
Otagiri M, Ui S, Takusagawa Y et al (2010) Structural basis for chiral substrate recognition by two 2,3-butanediol dehydrogenases. FEBS Lett 584:219–223. https://doi.org/10.1016/j.febslet.2009.11.068
Jongeneel CV, Bouvier J, Bairoch A (1989) A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett 242:211–214. https://doi.org/10.1016/0014-5793(89)80471-5
Brettrager EJ, Cuya SM, Tibbs ZE (2023) N-terminal domain of tyrosyl-DNA phosphodiesterase I regulates topoisomerase I-induced toxicity in cells. Sci Rep 13:1377. https://doi.org/10.1038/s41598-023-28564-6
Cala O, Guillière F, Krimm I (2014) NMR-based analysis of protein-ligand interactions. Anal Bioanal Chem 406:943–956. https://doi.org/10.1007/s00216-013-6931-0
Gohlke H, Klebe G (2002) Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew Chem Int Ed Engl 41:2644–2676. https://doi.org/10.1002/1521-3773(20020802)41:15%3c2644::AID-ANIE2644%3e3.0.CO;2-O
Rasul HO, Aziz BK, Ghafour DD et al (2021) In silico molecular docking and dynamic simulation of eugenol compounds against breast cancer. J Mol Model 28:17. https://doi.org/10.1007/s00894-021-05010-w
Meyer M, Wilson P, Schomburg D (1996) Hydrogen bonding and molecular surface shape complementarity as a basis for protein docking. J Mol Biol 264:199–210. https://doi.org/10.1006/jmbi.1996.0634
Mhatre S, Patravale V (2021) Drug repurposing of triazoles against mucormycosis using molecular docking: a short communication. Comput Biol Med 136:104722. https://doi.org/10.1016/j.compbiomed.2021.104722
Kwofie SK, Hanson G, Sasu H et al (2022) Molecular modelling and atomistic insights into the binding mechanism of MmpL3 Mtb. Chem Biodivers 19:e202200160. https://doi.org/10.1002/cbdv.202200160
Persson B, Hedlund J, Jörnvall H (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: the MDR superfamily. Cell Mol Life Sci 65:3879–3894. https://doi.org/10.1007/s00018-008-8587-z
Fu J, Wang Z, Chen T et al (2014) NADH plays the vital role for chiral pure D-(-)-2,3-butanediol production in Bacillus subtilis under limited oxygen conditions. Biotechnol Bioeng 111:2126–2131. https://doi.org/10.1002/bit.25265
Wang Y, San KY, Bennett GN (2013) Cofactor engineering for advancing chemical biotechnology. Curr Opin Biotechnol 24:994–999. https://doi.org/10.1016/j.copbio.2013.03.022
Bardhan M, Chowdhury J, Ganguly T (2011) Investigations on the interactions of aurintricarboxylic acid with bovine serum albumin: Steady state/time resolved spectroscopic and docking studies. J Photochem Photobiol B 102:11–19. https://doi.org/10.1016/j.jphotobiol.2010.08.011
Tang H, Huang L, Sun C et al (2020) Exploring the structure-activity relationship and interaction mechanism of flavonoids and α-glucosidase based on experimental analysis and molecular docking studies. Food Funct 11:3332–3350. https://doi.org/10.1039/c9fo02806d
Southall NT, Dill KA, Haymet DJ (2002) A view of the hydrophobic effect. J Phys Chem B 106:521–533. https://doi.org/10.1021/jp015514e
Abdullah SMS, Fatma S, Rabbani G et al (2017) A spectroscopic and molecular docking approach on the binding of tinzaparin sodium with human serum albumin. J Mol Struct 1127:283–288. https://doi.org/10.1016/j.molstruc.2016.07.108
Yamada R, Wakita K, Mitsui R et al (2017) Efficient production of 2,3-butanediol by recombinant Saccharomyces cerevisiae through modulation of gene expression by cocktail δ-integration. Bioresour Technol 245:1558–1566. https://doi.org/10.1016/j.biortech.2017.05.034
Yang T, Rao Z, Hu G et al (2015) Metabolic engineering of Bacillus subtilis for redistributing the carbon flux to 2,3-butanediol by manipulating NADH levels. Biotechnol Biofuels 8:129. https://doi.org/10.1186/s13068-015-0320-1
Pu Z, Ji F, Wang J et al (2017) Rational design of Meso-2,3-butanediol dehydrogenase by molecular dynamics simulation and experimental evaluations. FEBS Lett 591:3402–3413. https://doi.org/10.1002/1873-3468.12834
Acknowledgements
We acknowledge the Confederation of Indian Industry (CII) and Praj Industries Ltd., Pune, India, for the financial support to Sailee Sanjay Asolkar under the scheme “Prime Minister Fellowship for Doctoral research”. This work was funded by Praj Matrix-R and D Center (Division of Praj Industries Limited), Pune, India.
Author information
Authors and Affiliations
Contributions
SSA involved in data curation, investigation, methodology, formal analysis, validation, visualization, writing—original draft & editing. AM took part in silico investigation, methodology, formal analysis, validation, visualization, writing—original draft & editing. RK took part in silico investigation, methodology, formal analysis, validation, visualization, writing—review. AD involved in conceptualization, data curation, investigation, methodology, writing—review. AG involved in conceptualization, data curation, investigation, methodology, writing—review. PK involved in funding acquisition, project administration, resources.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Asolkar, S.S., Anju, M., Kumar, R. et al. In silico screening and validation of different dehydrogenases to produce 2,3-butanediol in Bacillus subtilis. Biotechnol Bioproc E 29, 271–290 (2024). https://doi.org/10.1007/s12257-024-00053-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-024-00053-1