Abstract
Purpose
Breast cancer with positive hormone receptor (HR) and human epidermal growth factor receptor-2 (HER2) is a special subgroup with different clinical features and survival, especially the endocrine therapy resistance. The main purpose of the study is to find the potential markers to predict the survival and endocrine therapy resistance of patients with HR+ /HER2+ breast cancer.
Methods
Surveillance, Epidemiology, and End Results (SEER) database was used to collect patients’ clinical information and tumor features including age, tumor size, grade, stage and long-term survival; the BioPortal for Cancer Genomics (https://cbioportal.org) was used to download the gene data for specific patient group; cluster analyses of gene expression were conducted through the DAVID Bioinformatics Resources 6.8 software.
Results
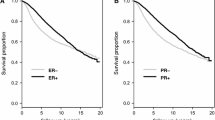
All of the included patients were diagnosed as HR positive breast cancer, but the PR positive rates were more common in HER2- group and also the ER+ /PR+ disease. Patients in HR+ /HER2+ group were more likely to present as stage III–IV and grade III disease. Among HR+ /HER2+ patients, 68.6% received chemotherapy, while only 28.9% in HR+ /HER2– group received chemotherapy (P < 0.0001). The survival of HR+ /HER2+ group was poorer. From TCGA database, series genes which were differed between HR+ /HER2+ and HR+ /HER2– were screened out that related to ERBB2 closely: IKZF3, LASP1, CDK12, MLLT6, and RARA. The first three candidate genes were associated with patients’ survival, especially in patients who received hormone therapies.
Conclusion
This study analyzed the clinical characteristics and survival of patients with HR+/HER2+ breast cancer as a special subgroup. ERBB2, IKZF3, LASP1, and CDK12 were the potential markers of the resistance of endocrine therapy, and they will provide new strategies for clinicians.




Similar content being viewed by others
References
Marcom PK, Isaacs C, Harris L, Wong ZW, Kommarreddy A, Novielli N, et al. The combination of letrozole and trastuzumab as first or second-line biological therapy produces durable responses in a subset of HER2 positive and ER positive advanced breast cancers. Breast Cancer Res Treat. 2007;102:43–9.
Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279.
Barton MK. Hormone receptor status affects locoregional control in HER2-positive breast cancer treated with trastuzumab. CA Cancer J Clin. 2012;62:281–2.
Chen Z, Wang Y, Warden C, Chen S. Cross-talk between ER and HER2 regulates c-MYC-mediated glutamine metabolism in aromatase inhibitor resistant breast cancer cells. J Steroid Biochem Mol Biol. 2015;149:118–27.
Cui J, Germer K, Wu T, Wang J, Luo J, Wang SC, et al. Cross-talk between HER2 and MED1 regulates tamoxifen resistance of human breast cancer cells. Cancer Res. 2012;72:5625–34.
Nahta R, O'Regan RM. Therapeutic implications of estrogen receptor signaling in HER2-positive breast cancers. Breast Cancer Res Treat. 2012;135:39–48.
Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem. 2001;276:9817–24.
Chen Z, Yang J, Li S, Lv M, Shen Y, Wang B, et al. Invasive lobular carcinoma of the breast: a special histological type compared with invasive ductal carcinoma. PLoS One. 2017;12:e0182397.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:11.
da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Xu M, Ren Z, Wang X, Comer A, Frank JA, Ke ZJ, et al. ErbB2 and p38gamma MAPK mediate alcohol-induced increase in breast cancer stem cells and metastasis. Mol Cancer. 2016;15:52.
Toomey S, Eustace AJ, Fay J, Sheehan KM, Carr A, Milewska M, et al. Impact of somatic PI3K pathway and ERBB family mutations on pathological complete response (pCR) in HER2-positive breast cancer patients who received neoadjuvant HER2-targeted therapies. Breast Cancer Rese BCR. 2017;19:87.
Synnott NC, Bauer MR, Madden S, Murray A, Klinger R, O'Donovan N, et al. Mutant p53 as a therapeutic target for the treatment of triple-negative breast cancer: preclinical investigation with the anti-p53 drug, PK11007. Cancer Lett. 2018;414:99–106.
Thaler S, Schmidt M, Robetawag S, Thiede G, Schad A, Sleeman JP. Proteasome inhibitors prevent bi-directional HER2/estrogen-receptor cross-talk leading to cell death in endocrine and lapatinib-resistant HER2+/ER+ breast cancer cells. Oncotarget. 2017;8:72281–30101.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–31.
Bhalla S, Chattree N. Value of clinico-pathological factors in prognosis of breast cancer patients. Indian J Pathol Microbiol. 1990;33:250–7.
Cao SS, Lu CT. Recent perspectives of breast cancer prognosis and predictive factors. Oncol Lett. 2016;12:3674–8.
Rasmussen BB, Regan MM, Lykkesfeldt AE, Dell'Orto P, Del Curto B, Henriksen KL, et al. Adjuvant letrozole versus tamoxifen according to centrally-assessed ERBB2 status for postmenopausal women with endocrine-responsive early breast cancer: supplementary results from the BIG 1–98 randomised trial. Lancet Oncol. 2008;9:23–8.
Dowsett M, Allred C, Knox J, Quinn E, Salter J, Wale C, et al. Relationship between quantitative estrogen and progesterone receptor expression and human epidermal growth factor receptor 2 (HER-2) status with recurrence in the arimidex, tamoxifen, alone or in combination trial. J Clin Oncol. 2008;26:1059–65.
Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–47.
Garcia-Becerra R, Santos N, Diaz L, Camacho J. Mechanisms of resistance to endocrine therapy in breast cancer: focus on signaling pathways, miRNAs and genetically based resistance. Int J Mol Sci. 2012;14:108–45.
Kaufman B, Mackey JR, Clemens MR, Bapsy PP, Vaid A, Wardley A, et al. Trastuzumab plus anastrozole versus anastrozole alone for the treatment of postmenopausal women with human epidermal growth factor receptor 2-positive, hormone receptor-positive metastatic breast cancer: results from the randomized phase III TAnDEM study. J Clin Oncol. 2009;27:5529–37.
Johnston S, Pippen J Jr, Pivot X, Lichinitser M, Sadeghi S, Dieras V, et al. Lapatinib combined with letrozole versus letrozole and placebo as first-line therapy for postmenopausal hormone receptor-positive metastatic breast cancer. J Clin Oncol. 2009;27:5538–46.
Kauraniemi P, Kuukasjarvi T, Sauter G, Kallioniemi A. Amplification of a 280-kilobase core region at the ERBB2 locus leads to activation of two hypothetical proteins in breast cancer. Am J Pathol. 2003;163:1979–84.
Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–19.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Tien JF, Mazloomian A, Cheng SG, Hughes CS, Chow CCT, Canapi LT, et al. CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion. Nucleic Acids Res. 2017;45:6698–716.
Harding E. Palbociclib for hormone receptor-positive breast cancer. Lancet Oncol. 2015;16:e318.
Johnson SF, Cruz C, Greifenberg AK, Dust S, Stover DG, Chi D, et al. CDK12 Inhibition reverses de novo and acquired parp inhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Rep. 2016;17:2367–81.
Tomasetto C, Regnier C, Moog-Lutz C, Mattei MG, Chenard MP, Lidereau R, et al. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics. 1995;28:367–76.
Grunewald TG, Kammerer U, Kapp M, Eck M, Dietl J, Butt E, et al. Nuclear localization and cytosolic overexpression of LASP-1 correlates with tumor size and nodal-positivity of human breast carcinoma. BMC Cancer. 2007;7:198.
Frietsch JJ, Grunewald TG, Jasper S, Kammerer U, Herterich S, Kapp M, et al. Nuclear localisation of LASP-1 correlates with poor long-term survival in female breast cancer. Br J Cancer. 2010;102:1645–53.
Benusiglio PR, Pharoah PD, Smith PL, Lesueur F, Conroy D, Luben RN, et al. HapMap-based study of the 17q21 ERBB2 amplicon in susceptibility to breast cancer. Br J Cancer. 2006;95:1689–95.
Acknowledgements
This work was supported by Natural Science Foundation of China (Grant number, 81802623).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that there are no conflicts of interest between authors.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not available.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, K., Quan, J., Yang, J. et al. The potential markers of endocrine resistance among HR+ /HER2+ breast cancer patients. Clin Transl Oncol 22, 576–584 (2020). https://doi.org/10.1007/s12094-019-02163-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-019-02163-2