Abstract
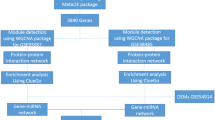
Schizophrenia (SCZ) is a polygenic, complex mental disorder of which a diagnosis is often made based on psychiatric history and clinical observation with few available objectives and detectable biomarkers. To identify co-expressed miRNA modules in schizophrenia patients and verify the possibility of using peripheral blood miRNAs as novel biomarkers, high-throughput sequencing was performed on 15 first-episode schizophrenia patients (FES) and 15 healthy controls (CTL). We found 79 differential expressed miRNAs (DEMs) in FES patients and three FES-related co-expression miRNA modules by miRNA-seq data standardized difference analysis and weighted gene co-expression network analysis (WGCNA). Then, 41 hub miRNAs were screened from the intersection of key modules and DEMs, among which miR-9-5p, miR-144-3p, miR-328-3p, and miR-4467 were selected for qRT-PCR verification in a larger sample (FES = 35, CTL = 60). The level of miR-9-5p in FES patients was downregulated, and miR-4467 was upregulated with better diagnostic performance (AUC = 0.719). The target genes of miR-9-5p engage in the biological processes (BP) such as body behaviour, neuronal differentiation regulation, nervous system development, and neurotrophin signaling pathways. Their hub target genes were also located, including NEDD4, EIF4G1, FBXL16, and FBXL3. Summarily, miR-9-5p and miR-4467 hold promise in blood diagnosis for SCZ, and miR-9-5p might affect the onset and development of SCZ through target regulation of neurodevelopment-related mRNAs. Our findings revealed the complex relationship between the miRNA co-expression network and FES, providing more verifiable biomarkers for SCZ early diagnosis and clues for the etiology of schizophrenia.








Similar content being viewed by others
Data Availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
References
Insel TR (2010) Rethinking schizophrenia. Nature 468(7321):187–193. https://doi.org/10.1038/nature09552
Charlson FJ, Ferrari AJ, Santomauro DF, Diminic S, Stockings E, Scott JG, McGrath JJ, Whiteford HA (2018) Global epidemiology and burden of schizophrenia: findings from the Global Burden of Disease Study 2016. Schizophr Bull 44(6):1195–1203. https://doi.org/10.1093/schbul/sby058
Laursen TM, Nordentoft M, Mortensen PB (2014) Excess early mortality in schizophrenia. Annu Rev Clin Psychol 10:425–448. https://doi.org/10.1146/annurev-clinpsy-032813-153657
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11(9):597–610. https://doi.org/10.1038/nrg2843
Lee YS, Dutta A (2009) MicroRNAs in cancer. Annu Rev Pathol 4:199–227. https://doi.org/10.1146/annurev.pathol.4.110807.092222
Zhou SS, Jin JP, Wang JQ, Zhang ZG, Freedman JH, Zheng Y, Cai L (2018) miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin 39(7):1073–1084. https://doi.org/10.1038/aps.2018.30
Rottiers V, Näär AM (2012) MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13(4):239–250. https://doi.org/10.1038/nrm3313
Miller BH, Wahlestedt C (2010) MicroRNA dysregulation in psychiatric disease. Brain Res 1338:89–99. https://doi.org/10.1016/j.brainres.2010.03.035
Sun E, Shi Y (2015) MicroRNAs: small molecules with big roles in neurodevelopment and diseases. Exp Neurol 268:46–53. https://doi.org/10.1016/j.expneurol.2014.08.005
Gallego JA, Gordon ML, Claycomb K, Bhatt M, Lencz T, Malhotra AK (2012) In vivo microRNA detection and quantitation in cerebrospinal fluid. J Mole Neurosci : MN 47(2):243–248. https://doi.org/10.1007/s12031-012-9731-7
Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM (2011) Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiat 69(2):188–193. https://doi.org/10.1016/j.biopsych.2010.09.039
Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J et al (2007) microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol 8(2):R27. https://doi.org/10.1186/gb-2007-8-2-r27
He K, Guo C, He L, Shi Y (2018) MiRNAs of peripheral blood as the biomarker of schizophrenia. Hereditas 155:9. https://doi.org/10.1186/s41065-017-0044-2
Wei H, Yuan Y, Liu S, Wang C, Yang F, Lu Z, Wang C, Deng H et al (2015) Detection of circulating miRNA levels in schizophrenia. Am J Psychiatry 172(11):1141–1147. https://doi.org/10.1176/appi.ajp.2015.14030273
Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, Cairns MJ (2012) Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry 17(8):827–840. https://doi.org/10.1038/mp.2011.78
Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1:54. https://doi.org/10.1186/1752-0509-1-54
Langfelder P, Luo R, Oldham MC, Horvath S (2011) Is my network module preserved and reproducible? PLoS Comput Biol 7(1):e1001057. https://doi.org/10.1371/journal.pcbi.1001057
Huang HY, Lin YC, Li J, Huang KY, Shrestha S, Hong HC, Tang Y, Chen YG et al (2020) miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res 48(D1):D148-d154. https://doi.org/10.1093/nar/gkz896
da Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT et al (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45(D1):D362-d368. https://doi.org/10.1093/nar/gkw937
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Luoni A, Riva MA (2016) MicroRNAs and psychiatric disorders: from aetiology to treatment. Pharmacol Ther 167:13–27. https://doi.org/10.1016/j.pharmthera.2016.07.006
Seo MS, Scarr E, Lai CY, Dean B (2014) Potential molecular and cellular mechanism of psychotropic drugs. Clin Psychopharmacol Neurosci 12(2):94–110. https://doi.org/10.9758/cpn.2014.12.2.94
Huang X, Bao C, Lv Q, Zhao J, Wang Y, Lang X, Li Z, Yi Z (2020) Sex difference in cognitive impairment in drug-free schizophrenia: association with miR-195 levels. Psychoneuroendocrinology 119:104748. https://doi.org/10.1016/j.psyneuen.2020.104748
Camkurt MA, Acar Ş, Coşkun S, Güneş M, Güneş S, Yılmaz MF, Görür A, Tamer L (2015) Comparison of plasma microRNA levels in drug naive, first episode depressed patients and healthy controls. J Psychiatr Res 69:67–71. https://doi.org/10.1016/j.jpsychires.2015.07.023
Wang X, Sundquist K, Hedelius A, Palmér K, Memon AA, Sundquist J (2015) Circulating microRNA-144-5p is associated with depressive disorders. Clin Epigenetics 7(1):69. https://doi.org/10.1186/s13148-015-0099-8
Wan Y, Liu Y, Wang X, Wu J, Liu K, Zhou J, Liu L, Zhang C (2015) Identification of differential microRNAs in cerebrospinal fluid and serum of patients with major depressive disorder. PLoS ONE 10(3):e0121975. https://doi.org/10.1371/journal.pone.0121975
Kichukova TM, Popov NT, Ivanov IS, Vachev TI (2017) Profiling of circulating serum microRNAs in children with autism spectrum disorder using stem-loop qRT-PCR assay. Folia Med 59(1):43–52. https://doi.org/10.1515/folmed-2017-0009
Ozkul Y, Taheri S, Bayram KK, Sener EF, Mehmetbeyoglu E, Öztop DB, Aybuga F, Tufan E et al (2020) A heritable profile of six miRNAs in autistic patients and mouse models. Sci Rep 10(1):9011. https://doi.org/10.1038/s41598-020-65847-8
Mor M, Nardone S, Sams DS, Elliott E (2015) Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex. Molecular autism 6:46. https://doi.org/10.1186/s13229-015-0040-1
Qin Z, Han X, Ran J, Guo S, Lv L (2021) Exercise-mediated alteration of miR-192–5p is associated with cognitive improvement in Alzheimer’s disease. Neuroimmunomodulation 29(1):36–43. https://doi.org/10.1159/000516928
Gámez-Valero A, Campdelacreu J, Vilas D, Ispierto L, Reñé R, Álvarez R, Armengol MP, Borràs FE et al (2019) Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Translational neurodegeneration 8:31. https://doi.org/10.1186/s40035-019-0169-5
Ghanbari M, Munshi ST, Ma B, Lendemeijer B, Bansal S, Adams HH, Wang W, Goth K et al (2019) A functional variant in the miR-142 promoter modulating its expression and conferring risk of Alzheimer disease. Hum Mutat 40(11):21312145. https://doi.org/10.1002/humu.23872
Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, Byrne M, Bernier F, Yanagimachi M et al (2013) Circulating miRNA biomarkers for Alzheimer’s disease. PLoS ONE 8(7):e69807. https://doi.org/10.1371/journal.pone.0069807
Van der Auwera S, Ameling S, Wittfeld K, d’HarcourtRowold E, Nauck M, Völzke H, Suhre K, Najafi-Shoushtari H et al (2019) Association of childhood traumatization and neuropsychiatric outcomes with altered plasma micro RNA-levels. Neuropsychopharmacol: official publication of the American College of Neuropsychopharmacology 44(12):2030–2037. https://doi.org/10.1038/s41386-019-0460-2
Zhou LT, Zhang J, Tan L, Huang HZ, Zhou Y, Liu ZQ, Lu Y, Zhu LQ, Yao C, Liu D (2021) Elevated levels of miR-144-3p induce cholinergic degeneration by impairing the maturation of NGF in Alzheimer’s disease. Front Cell Dev Biol 9:667412. https://doi.org/10.3389/fcell.2021.667412
Sun X, Wu Y, Gu M, Zhang Y (2014) miR-342-5p decreases ankyrin G levels in Alzheimer’s disease transgenic mouse models. Cell Rep 6(2):264–270. https://doi.org/10.1016/j.celrep.2013.12.028
Dakterzada F, David Benítez I, Targa A, Lladó A, Torres G, Romero L, de Gonzalo-Calvo D, Moncusí-Moix A, Tort-Merino A, Huerto R, Sánchez-de-la-Torre M, Barbé F, Piñol-Ripoll G (2021) Reduced levels of miR-342-5p in plasma are associated with worse cognitive evolution in patients with mild Alzheimer’s disease. Front Aging Neurosci 13:705989. https://doi.org/10.3389/fnagi.2021.705989
Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, Smalheiser NR (2015) Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS ONE 10(10):e0139233. https://doi.org/10.1371/journal.pone.0139233
Barbagallo C, Mostile G, Baglieri G, Giunta F, Luca A, Raciti L, Zappia M, Purrello M, Ragusa M, Nicoletti A (2020) Specific signatures of serum miRNAs as potential biomarkers to discriminate clinically similar neurodegenerative and vascular-related diseases. Cell Mol Neurobiol 40(4):531–546. https://doi.org/10.1007/s10571-019-00751-y
Su L, Wang C, Zheng C, Wei H, Song X (2018) A meta-analysis of public microarray data identifies biological regulatory networks in Parkinson’s disease. BMC Med Genomics 11(1):40. https://doi.org/10.1186/s12920-018-0357-7
Chen Z, Li Z, Jiang C, Jiang X, Zhang J (2019) MiR-92b-3p promotes neurite growth and functional recovery via the PTEN/AKT pathway in acute spinal cord injury. J Cell Physiol 234(12):23043–23052. https://doi.org/10.1002/jcp.28864
Xu L, Cao H, Xie Y, Zhang Y, Du M, Xu X, Ye R, Liu X (2019) Exosome-shuttled miR-92b-3p from ischemic preconditioned astrocytes protects neurons against oxygen and glucose deprivation. Brain Res 1717:66–73. https://doi.org/10.1016/j.brainres.2019.04.009
Liu X, Cui X, Guan G, Dong Y, Zhang Z (2020) microRNA-192-5p is involved in nerve repair in rats with peripheral nerve injury by regulating XIAP. Cell cycle (Georgetown, Tex) 19(3):326–338. https://doi.org/10.1080/15384101.2019.1710916
Lai WS, Xu B, Westphal KG, Paterlini M, Olivier B, Pavlidis P, Karayiorgou M, Gogos JA (2006) Akt1 deficiency affects neuronal morphology and predisposes to abnormalities in prefrontal cortex functioning. Proc Natl Acad Sci USA 103(45):16906–16911. https://doi.org/10.1073/pnas.0604994103
Chang CY, Chen YW, Wang TW, Lai WS (2016) Akting up in the GABA hypothesis of schizophrenia: Akt1 deficiency modulates GABAergic functions and hippocampus-dependent functions. Sci Rep 6:33095. https://doi.org/10.1038/srep33095
Huang CH, Pei JC, Luo DZ, Chen C, Chen YW, Lai WS (2014) Investigation of gene effects and epistatic interactions between Akt1 and neuregulin 1 in the regulation of behavioral phenotypes and social functions in genetic mouse models of schizophrenia. Front Behav Neurosci 8:455. https://doi.org/10.3389/fnbeh.2014.00455
Vogt D, Cho KKA, Lee AT, Sohal VS, Rubenstein JLR (2015) The parvalbumin/somatostatin ratio is increased in Pten mutant mice and by human PTEN ASD alleles. Cell Rep 11(6):944–956. https://doi.org/10.1016/j.celrep.2015.04.019
Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF (2006) Pten regulates neuronal arborization and social interaction in mice. Neuron 50(3):377–388. https://doi.org/10.1016/j.neuron.2006.03.023
Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, Palmer TD, Butte AJ, Brunet A (2009) FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5(5):527–539. https://doi.org/10.1016/j.stem.2009.09.014
Furukawa-Hibi Y, Kobayashi Y, Chen C, Motoyama N (2005) FOXO transcription factors in cell-cycle regulation and the response to oxidative stress. Antioxid Redox Signal 7(5–6):752–760. https://doi.org/10.1089/ars.2005.7.752
Gross DN, van den Heuvel AP, Birnbaum MJ (2008) The role of FoxO in the regulation of metabolism. Oncogene 27(16):2320–2336. https://doi.org/10.1038/onc.2008.25
Zhang X, Tang N, Hadden TJ (1813) Rishi AK (2011) Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 11:1978–1986. https://doi.org/10.1016/j.bbamcr.2011.03.010
Ermakov EA, Dmitrieva EM, Parshukova DA, Kazantseva DV, Vasilieva AR, Smirnova LP (2021) Oxidative stress-related mechanisms in schizophrenia pathogenesis and new treatment perspectives. Oxid Med Cell Longev 2021:8881770. https://doi.org/10.1155/2021/8881770
Orea-Soufi A, Dávila D, Salazar-Roa M, de Mar LM, Velasco G (2019) Phosphorylation of FOXO proteins as a key mechanism to regulate their activity. Methods Mol Biol 1890:51–59. https://doi.org/10.1007/978-1-4939-8900-3_5
Cantley LC (2002) The phosphoinositide 3-kinase pathway. Science 296(5573):1655–1657. https://doi.org/10.1126/science.296.5573.1655
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96(6):857–868. https://doi.org/10.1016/s0092-8674(00)80595-4
Schmidt MF, Gan ZY, Komander D, Dewson G (2021) Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities. Cell Death Differ 28(2):570–590. https://doi.org/10.1038/s41418-020-00706-7
Gu S, Cui F, Yin J, Fang C, Liu L (2021) Altered mRNA expression levels of autophagy- and apoptosis-related genes in the FOXO pathway in schizophrenia patients treated with olanzapine. Neurosci Lett 746:135669. https://doi.org/10.1016/j.neulet.2021.135669
Camkurt MA, Karababa F, Erdal ME, Bayazıt H, Kandemir SB, Ay ME, Kandemir H, Ay Ö, Çiçek E, Selek S, Taşdelen B (2016) Investigation of dysregulation of several microRNAs in peripheral blood of schizophrenia patients. Clin Psychopharmacol Neurosci 14(3):256–260. https://doi.org/10.9758/cpn.2016.14.3.256
Krichevsky AM, Sonntag KC, Isacson O, Kosik KS (2006) Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24(4):857–864. https://doi.org/10.1634/stemcells.2005-0441
Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S (2011) MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci 31(9):3407–3422. https://doi.org/10.1523/jneurosci.5085-10.2011
Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB (2010) MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6(4):323–335. https://doi.org/10.1016/j.stem.2010.02.015
Zhao C, Sun G, Li S, Shi Y (2009) A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol 16(4):365–371. https://doi.org/10.1038/nsmb.1576
Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, Papalopulu N (2012) microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat Neurosci 15(5):697–699. https://doi.org/10.1038/nn.3082
Otaegi G, Pollock A, Hong J, Sun T (2011) MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci 31(3):809–818. https://doi.org/10.1523/jneurosci.4330-10.2011
Hauberg ME, Roussos P, Grove J, Børglum AD, Mattheisen M (2016) Analyzing the role of microRNAs in schizophrenia in the context of common genetic risk variants. JAMA Psychiat 73(4):369–377. https://doi.org/10.1001/jamapsychiatry.2015.3018
Kranz TM, Goetz RR, Walsh-Messinger J, Goetz D, Antonius D, Dolgalev I, Heguy A, Seandel M, Malaspina D, Chao MV (2015) Rare variants in the neurotrophin signaling pathway implicated in schizophrenia risk. Schizophr Res 168(1–2):421–428. https://doi.org/10.1016/j.schres.2015.07.002
Su Y, Yang L, Li Z, Wang W, Xing M, Fang Y, Cheng Y, Lin GN, Cui D (2021) The interaction of ASAH1 and NGF gene involving in neurotrophin signaling pathway contributes to schizophrenia susceptibility and psychopathology. Prog Neuropsychopharmacol Biol Psychiatry 104:110015. https://doi.org/10.1016/j.pnpbp.2020.110015
Crider A, Pillai A (2017) Estrogen signaling as a therapeutic target in neurodevelopmental disorders. J Pharmacol Exp Ther 360(1):48–58. https://doi.org/10.1124/jpet.116.237412
Azcoitia I, Yague JG, Garcia-Segura LM (2011) Estradiol synthesis within the human brain. Neuroscience 191:139–147. https://doi.org/10.1016/j.neuroscience.2011.02.012
Cui J, Shen Y, Li R (2013) Estrogen synthesis and signaling pathways during aging: from periphery to brain. Trends Mol Med 19(3):197–209. https://doi.org/10.1016/j.molmed.2012.12.007
Denk J, Boelmans K, Siegismund C, Lassner D, Arlt S, Jahn H (2015) MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer’s disease. PLoS ONE 10(5):e0126423. https://doi.org/10.1371/journal.pone.0126423
Deng H, Wu Y, Jankovic J (2015) The EIF4G1 gene and Parkinson’s disease. Acta Neurol Scand 132(2):73–78. https://doi.org/10.1111/ane.12397
Han C, Cui K, Bi X, Wang L, Sun M, Yang L, Liu L (2019) Association between polymorphism of the NEDD4 gene and cognitive dysfunction of schizophrenia patients in Chinese Han population. BMC Psychiatry 19(1):405. https://doi.org/10.1186/s12888-019-2386-y
Smith AR, Smith RG, Pishva E, Hannon E, Roubroeks JAY, Burrage J, Troakes C, Al-Sarraj S, Sloan C, Mill J, van den Hove DL, Lunnon K (2019) Parallel profiling of DNA methylation and hydroxymethylation highlights neuropathology-associated epigenetic variation in Alzheimer’s disease. Clin Epigenetics 11(1):52. https://doi.org/10.1186/s13148-019-0636-y
Ansar M, Paracha SA, Serretti A, Sarwar MT, Khan J, Ranza E, Falconnet E, Iwaszkiewicz J, Shah SF, Qaisar AA, Santoni FA, Zoete V, Megarbane A, Ahmed J, Colombo R, Makrythanasis P, Antonarakis SE (2019) Biallelic variants in FBXL3 cause intellectual disability, delayed motor development and short stature. Hum Mol Genet 28(6):972–979. https://doi.org/10.1093/hmg/ddy406
Dong Z, Chen W, Chen C, Wang H, Cui W, Tan Z, Robinson H, Gao N, Luo B, Zhang L, Zhao K, Xiong WC, Mei L (2020) CUL3 Deficiency causes social deficits and anxiety-like behaviors by impairing excitation-inhibition balance through the promotion of cap-dependent translation. Neuron 105(3):475-490.e476. https://doi.org/10.1016/j.neuron.2019.10.035
Glynn D, Sizemore RJ, Morton AJ (2007) Early motor development is abnormal in complexin 1 knockout mice. Neurobiol Dis 25(3):483–495. https://doi.org/10.1016/j.nbd.2006.10.011
Glynn D, Drew CJ, Reim K, Brose N, Morton AJ (2005) Profound ataxia in complexin I knockout mice masks a complex phenotype that includes exploratory and habituation deficits. Hum Mol Genet 14(16):2369–2385. https://doi.org/10.1093/hmg/ddi239
Fujikawa R, Higuchi S, Nakatsuji M, Yasui M, Ikedo T, Nagata M, Yokode M, Minami M (2016) EP4 receptor-associated protein in microglia promotes inflammation in the brain. Am J Pathol 186(8):1982–1988. https://doi.org/10.1016/j.ajpath.2016.04.002
Fatemi SH, Folsom TD, Reutiman TJ, Novak J, Engel RH (2012) Comparative gene expression study of the chronic exposure to clozapine and haloperidol in rat frontal cortex. Schizophr Res 134(2–3):211–218. https://doi.org/10.1016/j.schres.2011.11.013
Nt P, Ds M, Mm N, In M, Ti V (2018) Investigation of circulating serum microRNA-328-3p and microRNA-3135a expression as promising novel biomarkers for autism spectrum disorder. Balkan J Med Genet 21(2):5–12. https://doi.org/10.2478/bjmg-2018-0026
Zhou Q, Luo L, Wang X, Li X (2019) Relationship between single nucleotide polymorphisms in the 3'UTR of amyloid precursor protein and risk of Alzheimer's disease and its mechanism. Biosci Rep 39(5):BSR20182485. https://doi.org/10.1042/bsr20182485
Everaert C, Luypaert M, Maag JLV, Cheng QX, Dinger ME, Hellemans J, Mestdagh P (2017) Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci Rep 7(1):1559. https://doi.org/10.1038/s41598-017-01617-3
Acknowledgements
We would like to thank the professional help provided by the psychiatrists in the study. We are very grateful to all the participants who volunteered to participate in this study.
Funding
This work was supported by the National Natural Science Foundation of China [81673253] and the Jilin Provincial Ministry of Education S & T Project [JJKH20190091KJ].
Author information
Authors and Affiliations
Contributions
QY and MDJ conceived and designed the study. XJZ and YYS did the material preparation. Data analysis and experiment were performed by MDJ and ZJL. MDJ and XWL wrote the first draft of the manuscript. NNJ and YEL collected the data. LZA, GYH, XYC, MTX, and YQY revised the graphs. All authors commented on the previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
This study was performed in line with the principles of the Declaration of Helsinki. The study was approved by the Ethics Committee of the School of Public Health of Jilin University.
Consent to Participate
Informed consent was obtained from all individual participants or their guardians included in the study.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jin, M., Zhu, X., Sun, Y. et al. Identification of Peripheral Blood miRNA Biomarkers in First-Episode Drug-Free Schizophrenia Patients Using Bioinformatics Strategy. Mol Neurobiol 59, 4730–4746 (2022). https://doi.org/10.1007/s12035-022-02878-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-02878-4