Abstract
This study aimed to investigate the association between selected variants of genes related to dopamine metabolism pathways and the risk of and progression of Parkinson’s disease (PD). This prospective cohort study was conducted in one academic teaching hospital. The study was conducted on 126 patients diagnosed with idiopathic Parkinson’s disease. Blood samples were collected to conduct a genotyping of MAOB, DRD1, DRD2, and DDC genes. Genotype and allele frequencies of MAOB (rs1799836) variants were not associated with the course of PD. Genotype and allele frequencies of DRD2 (rs2283265) variants were associated with risk of dementia (p = 0.001) and resulted in parts II and III of the UPDRS scale (p = 0.001). Genotype and allele frequencies of DRD2 (rs1076560) variants were associated with risk of dementia (p = 0.001) and resulted in parts II and III of the UPDRS scale (p = 0.001). Genotype and allele frequencies of DDC (rs921451) variants were not associated with the course of PD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parkinson’s disease (PD) is a chronic and progressive neurodegenerative disorder that affects over 4 million people worldwide (Kalia and Lang 2015). PD is the second, after Alzheimer’s disease, the most common neurodegenerative disorder. Clinically, it is characterized by bradykinesia and at least one of the following symptoms: resting tremor, rigidity, and postural instability (Cacabelos 2017). The research has shown that genetic factors play a pivotal role in developing PD, but especially with youth-onset, but the exact etiology of this disorder is still very elusive (Post et al. 2020). However, the pathogenesis of PD is quite complex and still not fully understood. Previous research suggests that dysfunction of dopaminergic neurotransmission is involved in the pathogenesis of PD (Poewe et al. 2017). Dopaminergic signaling is crucial for motor planning and mental function. Available research suggests the existence of the associations between single nucleotide polymorphisms (SNPs) in different genes involved in neurotransmitter metabolism pathways (Connolly and Lang 2014). However, the exact role of polymorphic variants in dopaminergic neurotransmission regulation is yet to be determined. It is well known that there is a high level of genetic heterogeneity between different populations, and it is necessary to conduct multiple studies to identify the associations between genetic variations in genes that may be related to PD pathophysiology (Pastor 2012).
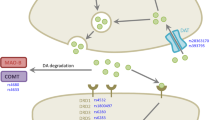
Monoamine oxidase B (MAOB) is an enzyme-containing flavin-adenine dinucleotide that plays a vital role in the inactivation of neurotransmitters especially by deamination of dopamine. The MAOB gene is located at chromosome Xp11.23. A644G single nucleotide polymorphism (rs1799836), located in exon 13 of the MAOB, is a known functional polymorphism (Kiyohara et al. 2011). A644G was shown to be associated with altered enzyme activity (Liu et al. 2014). The G allele of MAOB A644G polymorphism was associated with lower brain MAOB activity related to PD (Kakinuma et al. 2020).
Dopamine D2 receptor (DRD2) is a G-protein-coupled, and it is mainly expressed in the striatum. The DRD2 gene is located on chromosome 11q23.2. It encodes two molecularly distinct isoforms [the D2 long (D2L) and the D2 short (D2S)] with distinct functions (Gluskin and Mickey 2016; Dubovyk and Manahan-Vaughan 2019). Zhang et al. characterized two SNPs within DRD2, one in intron 5 rs2283265 and the second in intron 6 rs1076560. Both of them were demonstrated to be responsible for splicing. It has been shown that the T allele of both intronic SNPs caused shift splicing from D2S to D2L, which was associated with the changes in glutamate release (Zhang et al. 2007). Previous research has shown an association between these DRD2 polymorphisms and PD phenotypes related to motor fluctuations and dyskinesias (Rieck et al. 2012), whereas the other researchers have shown no associations (Kaiser et al. 2003; Kaplan et al. 2014; Lee et al. 2011). DOPA decarboxylase (DDC) is a pyridoxal 5′-phosphate-reliant enzyme encoded by the DDC gene located on chromosome 7p12.1. This enzyme can facilitate the synthesis of critical neuro-active biogenic amines in the brain (Zhu et al. 2013). Moreover, it was shown that the activity of this enzyme plays a very essential role in central nervous system physiology. Very rare that loss-of-function mutations in the DDC gene lead to a devastating neuro-developmental syndrome known as DDC deficiency (OMIM#608,643). This syndrome, resulting from DDC enzymatic insufficiency, causes severe autonomic, motor, and cognitive impairments which may manifest in early life (Brun et al. 2010). In the context of PD, no studies were showing the association between the genetic variation of the DDC gene with PD development and progression.
This study aimed to investigate the association between selected variants of genes related to dopamine metabolism pathways, representative, functional polymorphisms (rs1799836 of MAOB; rs5326 of DRD1; rs2283265, rs1800497, rs1801028, rs1799732, rs1076560 of DRD2, and rs1065852 of DDC), and the risk of and progression of Parkinson`s disease. The hypothesis was that the specific variants in selected genes involved in the dopamine metabolic pathway were associated with slowing the progression of Parkinson’s disease and improved the effectiveness of levodopa treatment.
Results
Study Group
The study was conducted on 126 patients diagnosed with idiopathic Parkinson’s disease. 49 (38.9%) women and 77 (61.1%) men. The median age was 67 years. The control group consisted of 94 individuals (34 women and 60 men) 35–91 years of age. The ethnicity of all patients was Caucasian. In half of the subject’s PD duration was about 9 years. Dementia (MMSE < 24 points) occurred in 23.4% of the patients. The median UPDRS score was 32 points. The complications of levodopa treatment (motor fluctuation, dyskinesia, dystonic movements) were observed in 68.8% of the patients. Deep brain stimulation was applied in 17.5% of the subjects. The distribution of selected characteristics among PD patients is shown in Table 1.
Genotype and Allele Frequencies of MAOB (rs1799836) Variant
First, we genotyped the MAOB (rs1799836) SNP in all DNA samples using the TaqMan assay, achieving a success rate of 100%. About 42.3% of the subjects had T/T genotype, 39.0% had C/C genotype, and 18.7% had T/C genotype within the rs1799836 polymorphism of the MAOB gene. In this case, we did not observe any significant differences in the frequency distribution of rs1799836 between the total case and control groups. There was no significant relationship between genotype variant and incidence of dementia, motor fluctuations, and application of deep brain stimulation in patients: p = 0.116, p = 0.497, and p = 0.654, respectively. The p-value for the relationship between genotype variant and patients’ results in parts II and III of the UPDRS scale was 0.326.
Genotype and Allele Frequencies of DRD2 (rs2283265) Variant
The vast majority of patients with PD (75.0%) had A/A genotype, 13.3% had C/C genotype, and 11.7% had A/C genotype within the rs2283265 polymorphism of DRD2 gene (Table 2). There was a significant relationship between genotype variants and incidence of dementia (p = 0.001). The mean ± SD age of disease onset in patients with dementia was 57 ± 11, while in patients without dementia, it was 55 ± 12. The mean ± SD duration of the disease in the PD subjects with dementia was 18.3 ± 5.8, while in patients without dementia, it was 13.4 ± 4.7. We excluded the patients with dementia with Lewy bodies from the statistical analyses when correlating rs2283265 frequency with dementia. The mean ± SD score in MMSE in patients with dementia was 19.1 ± 6.8, while in patients without dementia, it was 25.6 ± 2.8. All the patients with PD without or with dementia met the clinical diagnostic criteria for PD. There was no significant association between rs2283265 variant and treatment complications or application of deep brain stimulation: p = 0.532 and p = 0.105, respectively. Only 3.4% of the patients with A/A genotype had dementia. On the other hand, all the patients with C/C genotype had Parkinson’s disease with dementia, and 71.4% of the patients with the A/C genotype had dementia. The relationship between genotype variant and patients’ results in parts II and III of the UPDRS scale was found (p = 0.001) (Table 3). Significant differences were found between A/A and C/C genotypes (Table 4). The median score of the patients with A/A genotype was 29 points, and that of the patients with C/C genotype was 50.5 points in parts II and III of the UPDRS scale. Moreover, a difference between A/A and A/C genotypes was observed. Half of the patients with A/A genotype obtained 29 points in parts II and III of the UPDRS scale, while half of those with A/C genotype obtained 64 points. There were no statistical differences (χ2 = 1263; df = 2; p = 0,532) when correlating the rs2283265 with the effectiveness of levodopa treatment.
Genotype and Allele Frequencies of DRD2 (rs1076560) Variant
Overall, 49.6% of the subjects had an A/C genotype. Almost the same percentage of patients (48.8%) had C/C genotype, while only 1.6% had A/A genotype within the rs1076560 polymorphism of DRD2 gene (Table 5). There was a relationship between genotype variants and incidence of dementia (p < 0.001). For the other tested variables — motor fluctuations and application of deep brain stimulation, the p-values were 0.655 and 0.605, respectively. None of the patients with the A/A genotype had dementia. In the A/C genotype group patients, 45.0% had a form of PD with dementia. Among the patients with C/C genotype, only 3.4% had dementia. Due to the insufficient number of subjects with A/A genotype (2), it was necessary to exclude them from assessing the relationship between genotype variants and the results of the UPDRS scale. There was a relationship between genotype variants and patients’ results in parts II and III of the UPDRS scale for A/C and C/C genotypes (p < 0.001). The average results of the patients with A/C and C/C genotypes in this scale were: 43.3 and 26.2 points, respectively (Table 6). The statistical significant differences were not observed (χ2 = 0.845; df = 2; p = 0.655) when correlating the frequency of rs1076560 with the effectiveness of levodopa treatment.
Genotype and Allele Frequencies of DDC (rs921451)
Overall, 48.4% of the patients had C/T genotype, 33.0% had T/T genotype, and 18.7% had C/C genotype within the rs921451 polymorphism of the DDC gene. There was no significant relationship between genotype variant and incidence of dementia, motor fluctuations, and application of deep brain stimulation: p = 0.796, p = 0.205, and p = 0.394, respectively. The p-value for the relationship between genotype variant and patients’ results in parts II and III of the UPDRS scale was 0.239. We did not observe the statistical significant correlation (χ2 = 3166; df = 2; p = 0.205) between the rs921451 and the side effects of levodopa treatment.
Discussion
This study investigated the associations between polymorphisms within the MAOB (rs1799836), DRD2 (rs228365 and rs1076560), DDC (rs921451) genes and phenotypes in PD patients treated with levodopa. We hypothesized that the selected polymorphisms are associated with the progression of the PD and levodopa treatment complication. Understanding the etiology of PD and providing a successful treatment remains a challenge. Identifying potential predictors of the treatment outcomes paves the way for more personalization and an individual approach to the treatment. The MAOB rs1799836 variant was not associated with the PD phenotype. We did not find any statistically significant associations of MAOB variant with the occurrence of the PD in the Polish population. There was no significant relationship between motor fluctuation, side-effects of the levodopa treatment, the onset of dementia or deep-brain stimulation treatment, and MAOB polymorphism.
Kiyohara et al. reported that polymorphism rs1799836 of the MAOB gene might play an important role in PD susceptibility in the Japanese population (Kiyohara et al. 2011). This association was later confirmed in a meta-analysis by Sun et al. published in 2016 (Sun et al. 2014). Hao et al. verified the genotype frequencies of MAO-B rs1799836 A/AA, AG, G/GG in the Chinese PD patients’ population, 74.4%, 14.1%, and 11.5%, respectively (Hao et al. 2015). A study by Moreau et al. showed no significant associations between the response to L-dopa during PD treatment and MAOB rs1799836 variant (Moreau et al. 2015). A more recent study identified the rs1799836 polymorphism as a potential predictor of putaminal dopamine turnover in early PD.
Additionally, the MAOB TT allele was linked to high enzyme activity leading to higher intrinsic dopamine turnover, which has been demonstrated to constitute a risk factor for motor complications (Löhle et al. 2018). Sampaio et al. reported that patients carrying MAO-B (rs1799836) A and AA genotypes are prone to levodopa-induced-dyskinesia (Sampaio et al. 2018).
Presented results demonstrate that the CC genotype of DRD2 rs228365 was associated with dementia compared to the AA genotype. The study also suggests the CC genotype of DRD2 rs228365 as a genetic marker of more severe impairments in PD patients. The patients with this genotype had the UPDRS part II and III score of above 50 compared to the AA genotype (with a UPDRS part II and III score of below 29). The AA genotype was associated with a lower progression of PD. There are very few studies investigating the role of DRD2 rs228365 polymorphism in patients suffering from PD. There was no correlation between the rs2283265 polymorphism in the DRD2 gene and levodopa treatment and the need for deep-brain stimulation treatment. Authors of similar studies have shown the effect of rs2283265 on the treatment of PD patients as they were able to demonstrate that the polymorphism influenced the development of dyskinesias.
According to the results obtained by Rieck et al., a haplotype (TTCTA) derived from rs2283265 polymorphism in the DRD2 gene region is associated with dyskinesia during levodopa treatment, which results from the reduced expression of the DRD2 gene (Rieck et al. 2012). This results from the inhibition of negative feedback and lower control over dopamine release. Masellis et al. reported that single nucleotide polymorphisms rs2283265 in the DRD2 were found to be significantly associated with a favorable peak response to rasagiline at 12 weeks in early Parkinson’s disease (Masellis et al. 2016). Previous studies also identified associations between this polymorphism, other genetic variants, and severe cocaine abuse (Sullivan et al. 2013; Stolf et al. 2019). There are also reports highlighting the potential role of this polymorphism in developing various psychiatric disorders, including schizophrenia, ADHD, or others from the autism spectrum (Glatt et al. 2009; Gadow et al. 2014).
According to our results, the A/C genotype in rs1076560 polymorphism of the DRD2 gene is associated with dementia. It is essential to mention that the AA genotype was underrepresented in our study group. The patients with the A/C genotype had also significantly higher mean scores in parts II and III of the UPDRS scale.
An article by Miller et al. presented results suggesting a role of rs1076560 DRD2 polymorphism in predicting Parkinson’s disease gait impairment and medication responsiveness of specific gait functions. The authors explained that observed outcomes might result from reduced striatal D2 receptor expression (T allele carriers), which induces gait dysfunction compared to homozygous patients for the G allele (Miller et al. 2018). Masellis et al. identified rs1076560 polymorphism in the gene DRD2 (besides rs2283265) as associated with the peak clinical response to rasagiline (Masellis et al. 2016).
The DDC rs921451 variant was not associated with the PD phenotype. There were no statistically significant associations between DDC variants and PD treatment in the studied group. Also, DDC rs921451 polymorphism did not influence side-effects of the levodopa treatment, the onset of dementia during PD, or deep-brain stimulation treatment.
The rs921451 variant of the DDC gene was reported by Devos et al. to influence the motor response to L-dopa but do not significantly change peripheral pharmacokinetic parameters for L-dopa and dopamine (Devos et al. 2014). However, results reported by Moreau et al. presented no significant association between the rs921451 variant of the DDC gene and the response to L-dopa (Moreau et al. 2015). When the role of the rs921451 polymorphism of the DDC in striatal dopamine turnover in de novo PD was investigated, the authors did not report significant findings (Löhle et al. 2018). Publication by Redenšek et al. reported that carriers of at least one DDC rs921451 C allele had higher odds for developing orthostatic hypotension during dopaminergic treatment of the PD (Redenšek et al. 2019).
There are several limitations to this study. It was conducted in one center and included patients from one country. The study group was relatively homogenous in terms of the ethnicity of patients. Therefore, the generalizability of the results obtained is yet to be determined. Additionally, PD includes multiple types and subtypes. Their division is based on scoring patterns for baseline motor, cognitive, and psychiatric measures. Unfortunately, we could not recruit patients with different types of PD to the study group. It may further limit the generalizability of the results over differently manifested types of PD. There is a risk that investigated polymorphisms play a different role in different clinical manifestations.
Materials and Methods
Study Design
This prospective cohort study was conducted in one academic hospital between November 2018 and November 2020. A neurologist expert of movement disorders identified all the patients with PD (based on the UK Parkinson’s Disease Society Brain Bank Criteria for clinically probable disease). All the PD patients underwent the following clinical evaluation: Mini-Mental State Examination (MMSE), motor examination according to the Unified Parkinson’s Disease Rating Scale (UPDRS, part II and part III), and Hoehn and Yahr Staging (H&Y). ON–OFF motor fluctuations, dyskinesia, dystonia, off stage were monitored using the patients’ diary. All the subjects were invited to provide a blood sample for genetic assessment. The patients in the control group had no previous diagnosis of neurodegenerative or malignant disease or family history of these disorders.
TaqMan SNP Genotyping
From all the individuals, we collected blood samples using EDTA tubes. Genomic DNA (gDNA) was extracted from whole blood using silica-based spin columns such as the QIAam DNA Blood Mini Kit from Qiagen (Valencia, CA). The quality of gDNA was assessed using NanoDrop ND-1000 spectrophotometer from NanoDrop Technologies (Wilmington, DE, USA), and only high-quality gDNA was used for genotyping. DNA samples were then stored at –20 °C until genotype analysis. The genotyping was carried out with ready-to-order Custom TaqMan SNP genotyping assays (from Applied Biosystems, Foster City, CA). We performed the genotyping of only the key SNPs utilized to identify MAOB (rs1799836, assay ID C__8878790_10), DRD1 (rs4532, C__1011777_10), DRD2 (rs2283265, assay ID C__16070796_10, rs1800497, C__7486676_10, rs1801028, C__10725_20, rs1799732, C__33641686_10, rs1076560, assay ID C__2278888_10) and DDC (rs1065852, assay ID C__8320238_10; from Applied Biosystems). The assay used in the study included forward and reverse PCR primers for selected SNPs and two differently labeled TaqMan minor groove binder (MGB) probes. The biallelic SNP was located in the middle third of the probe. Each allele-specific MGB probe was labeled with a fluorescent reporter dye (either a FAM or a VIC reporter molecule) and was attached to a fluorescence quencher. For the intact MGB probe, the reporter dye was quenched. We used 20 ng of gDNA for amplification as per manufacturer’s directions scaled to a total volume of 25 µl. During PCR, the 50-nuclease activity of Taq DNA polymerase cleaved the reporter dye (FAM or VIC) from an MGB probe that was completely hybridized to the DNA strand. The PCR was performed according to the manufacturer’s instructions provided by Applied Biosystems. The PCR thermal cycling was as follows: initial denaturing at 95 °C for 30 s; 40 cycles of 92 °C for 5 s and 60 °C for 20 s. Post-amplification products were analyzed on an Applied Biosystems ViiA 7 Real-Time PCR System, and genotype calls were determined manually by a comparison to no template controls. An increase in either FAM or VIC dye fluorescence indicated homozygosity for FAM- or VIC-specific alleles (X:X or Y:Y), and an increase in the fluorescence of both dyes indicated heterozygosity (X:Y). The two colors (FAM or VIC) were detected using ViiA 7 Realt-Time PCR System. Genotype calls were assessed with Applied Biosystems TaqMan Genotyper Software.
Statistical Analyses
Data were analyzed using the Statistica software version 10. The genotype distribution and allele frequency of selected SNPs of MAOB, DRD1, DRD2, and DDC gene in the control and PD group were compared using chi-squared test. Bonferroni’s post hoc test was used to determine the differences between the groups. We assessed Hardy–Weinberg equilibrium via a goodness-of-fit χ2-test Kruskal–Wallis test R to compare the observed and expected genotype results in examined groups. A p-value ≤ 0.005 was considered statistically significant. At first, the genotypic and allelic distribution in both (study and control group) were analyzed, and only the statistically significant results were selected to the further analysis in which the genotypic and allelic distribution in the group of Parkinson disease patients with different disease progression and with or without levodopa-induced dyskinesia were analyzed.
Conclusions
Our data provide a solid from which we can work towards the personalization of PD treatment since it may help identify fragile PD patients who would benefit from a less aggressive dopaminergic treatment. All of the receptors are important and involved in the dopamine pathway. Further studies with a larger population will help clarify the interpretation of these data.
Data Availability
Data will be made available upon reasonable request.
References
Brun L, Ngu LH, Keng WT, Ch'Ng GS, Choy YS, Hwu WL, Lee WT, Willemsen MA, Verbeek MM, Wassenberg T, Regal L (2010) Clinical and biochemical features of aromatic L-amino acid decarboxylase deficiency. Neurology 75:64–71. https://doi.org/10.1212/WNL.0b013e3181e620ae
Cacabelos R (2017) Parkinson’s disease: from pathogenesis to pharmacogenomics. Int J Mol Sci 2017:18. https://doi.org/10.3390/ijms18030551
Connolly BS, Lang AE (2014) Pharmacological treatment of Parkinson disease: a review. JAMA 311:1670–1683. https://doi.org/10.1001/jama.2014.3654
Devos D, Lejeune S, Cormier-Dequaire F, Tahiri K, Charbonnier-Beaupel F, Rouaix N, Duhamel A, Sablonnière B, Bonnet A-M, Bonnet C et al (2014) Dopa-decarboxylase gene polymorphisms affect the motor response to L-dopa in Parkinson’s disease. Parkinsonism Relat Disord 20:170–175. https://doi.org/10.1016/j.parkreldis.2013.10.017
Dubovyk V, Manahan-Vaughan D (2019) Gradient of expression of dopamine D2 receptors along the dorso-ventral axis of the hippocampus. Front Synaptic Neurosci 11:28. https://doi.org/10.3389/fnsyn.2019.00028
Gadow KD, Pinsonneault JK, Perlman G, Sadee W (2014) Association of dopamine gene variants, emotion dysregulation and ADHD in autism spectrum disorder. Res Dev Disabil 35:1658–1665. https://doi.org/10.1016/j.ridd.2014.04.007
Glatt SJ, Faraone SV, Lasky-Su JA, Kanazawa T, Hwu H-G, Tsuang MT (2009) Family-based association testing strongly implicates DRD2 as a risk gene for schizophrenia in Han Chinese from Taiwan. Mol Psychiatry 14:885–893. https://doi.org/10.1038/mp.2008.30
Gluskin BS, Mickey BJ (2016) Genetic variation and dopamine D2 receptor availability: a systematic review and meta-analysis of human in vivo molecular imaging studies. Transl Psychiatry 6:e747. https://doi.org/10.1038/tp.2016.22
Hao H, Shao M, An J, Chen C, Feng X, Xie S, Gu Z, Chen B (2015) [Polymorphisms of catechol-O-methyltransferase and monoamine oxidase B genes among Chinese patients with Parkinson’s disease]. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi = Chinese. J Med Genet 32:1–5. https://doi.org/10.3760/cma.j.issn.1003-9406.2015.01.001
Kaiser R, Hofer A, Grapengiesser A, Gasser T, Kupsch A, Roots I, Brockmöller J (2003) L -dopa-induced adverse effects in PD and dopamine transporter gene polymorphism. Neurology 60:1750–1755. https://doi.org/10.1212/01.wnl.0000068009.32067.a1
Kakinuma S, Beppu M, Sawai S, Nakayama A, Hirano S, Yamanaka Y, Yamamoto T, Masafumi C, Aisihaer X, Aersilan A, Gao Y (2020) Monoamine oxidase B rs1799836 G allele polymorphism is a risk factor for early development of levodopa-induced dyskinesia in Parkinson’s disease. eNeurologicalSci 19:100239. https://doi.org/10.1016/j.ensci.2020.100239
Kalia LV, Lang AE (2015) Parkinson’s disease. Lancet (london, England) 386:896–912. https://doi.org/10.1016/S0140-6736(14)61393-3
Kaplan N, Vituri A, Korczyn AD, Cohen OS, Inzelberg R, Yahalom G, Kozlova E, Milgrom R, Laitman Y, Friedman E et al (2014) Sequence variants in SLC6A3, DRD2, and BDNF genes and time to levodopa-induced dyskinesias in Parkinson’s disease. J Mol Neurosci 53:183–188. https://doi.org/10.1007/s12031-014-0276-9
Kiyohara C, Miyake Y, Koyanagi M, Fujimoto T, Shirasawa S, Tanaka K, Fukushima W, Sasaki S, Tsuboi Y, Yamada T et al (2011) Genetic polymorphisms involved in dopaminergic neurotransmission and risk for Parkinson’s disease in a Japanese population. BMC Neurol 11:89. https://doi.org/10.1186/1471-2377-11-89
Lee JY, Cho J, Lee E-K, Park SS, Jeon BS (2011) Differential genetic susceptibility in diphasic and peak-dose dyskinesias in Parkinson’s disease. Mov Disord 26:73–79. https://doi.org/10.1002/mds.23400
Liu Y, Wang Z, Zhang B (2014) The relationship between monoamine oxidase B (MAOB) A644G polymorphism and Parkinson disease risk: a meta-analysis. Ann Saudi Med 34:12–17. https://doi.org/10.5144/0256-4947.2014.12
Löhle M, Mangone G, Wolz M, Beuthien-Baumann B, Oehme L, van den Hoff J, Kotzerke J, Reichmann H, Corvol J-C, Storch A (2018) Functional monoamine oxidase B gene intron 13 polymorphism predicts putaminal dopamine turnover in de novo Parkinson’s disease. Mov Disord 33:1496–1501. https://doi.org/10.1002/mds.27466
Masellis M, Collinson S, Freeman N, Tampakeras M, Levy J, Tchelet A, Eyal E, Berkovich E, Eliaz RE, Abler V et al (2016) Dopamine D2 receptor gene variants and response to rasagiline in early Parkinson’s disease: a pharmacogenetic study. Brain 139:2050–2062. https://doi.org/10.1093/brain/aww109
Miller NS, Chou KL, Bohnen NI, Müller MLTM, Seidler RD (2018) Dopaminergic polymorphisms associated with medication responsiveness of gait in Parkinson’s disease. Parkinsonism Relat Disord 48:54–60. https://doi.org/10.1016/j.parkreldis.2017.12.010
Moreau C, Meguig S, Corvol J-C, Labreuche J, Vasseur F, Duhamel A, Delval A, Bardyn T, Devedjian J-C, Rouaix N et al (2015) Polymorphism of the dopamine transporter type 1 gene modifies the treatment response in Parkinson’s disease. Brain 138:1271–1283. https://doi.org/10.1093/brain/awv063
Pastor P (2012) Genetic heterogeneity in Parkinson disease. Neurology 79:619–620. https://doi.org/10.1212/WNL.0b013e318264e3d2
Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, Schrag A-E, Lang AE (2017) Parkinson Disease Nat Rev Dis Prim 3:17013. https://doi.org/10.1038/nrdp.2017.13
Post B, van den Heuvel L, van Prooije T, van Ruissen X, van de Warrenburg B, Nonnekes J (2020) Young onset Parkinson’s disease: a modern and tailored approach. J Parkinsons Dis 10:S29–S36. https://doi.org/10.3233/JPD-202135
Redenšek S, Flisar D, Kojović M, Gregorič Kramberger M, Georgiev D, Pirtošek Z, Trošt M, Dolžan V (2019) Dopaminergic pathway genes influence adverse events related to dopaminergic treatment in Parkinson’s disease. Front Pharmacol 10:8. https://doi.org/10.3389/fphar.2019.00008
Rieck M, Schumacher-Schuh AF, Altmann V, Francisconi CL, Fagundes PT, Monte TL, Callegari-Jacques SM, Rieder CR, Hutz MH (2012) DRD2 haplotype is associated with dyskinesia induced by levodopa therapy in Parkinson’s disease patients. Pharmacogenomics 13:1701–1710. https://doi.org/10.2217/pgs.12.149
Sampaio TF, Dos Santos EUD, de Lima GDC, Dos Anjos RSG, da Silva RC, Asano AGC, Asano NMJ, Crovella S, de Souza PRE (2018) MAO-B and COMT genetic variations associated with levodopa treatment response in patients With Parkinson’s disease. J Clin Pharmacol 58:920–926. https://doi.org/10.1002/jcph.1096
Stolf AR, Cupertino RB, Müller D, Sanvicente-Vieira B, Roman T, Vitola ES, Grevet EH, von Diemen L, Kessler FHP, Grassi-Oliveira R et al (2019) Effects of DRD2 splicing-regulatory polymorphism and DRD4 48 bp VNTR on crack cocaine addiction. J Neural Transm 126:193–199. https://doi.org/10.1007/s00702-018-1946-5
Sun Y-X, Wang X-H, Xu A-H, Zhao J-H (2014) Functional polymorphisms of the MAO gene with Parkinson disease susceptibility: a meta-analysis. J Neurol Sci 345:97–105. https://doi.org/10.1016/j.jns.2014.07.016
Sullivan D, Pinsonneault JK, Papp AC, Zhu H, Lemeshow S, Mash DC, Sadee W (2013) Dopamine transporter DAT and receptor DRD2 variants affect risk of lethal cocaine abuse: a gene-gene-environment interaction. Transl Psychiatry 3:e222–e228. https://doi.org/10.1038/tp.2012.146
Zhang Y, Bertolino A, Fazio L, Blasi G, Rampino A, Romano R, Lee M-LT, Xiao T, Papp A, Wang D et al (2007) Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci U S A 104:20552–20557. https://doi.org/10.1073/pnas.0707106104
Zhu B, Chen C, Moyzis RK, Dong Q, Chen C, He Q, Li J, Li J, Lei X, Lin C (2013) The DOPA decarboxylase (DDC) gene is associated with alerting attention. Prog Neuropsychopharmacol Biol Psychiatry 43:140–145. https://doi.org/10.1016/j.pnpbp.2012.12.020
Funding
This work was supported by the Institutional funding of the Jagiellonian University Medical College and Ministry of Science and Higher Education (Grant No: K/PBP/000318). The manuscript was reviewed by McGregor Language School.
Author information
Authors and Affiliations
Contributions
Conceptualization, B. Z. and R. M.; methodology, B. Z. and S. C.; software, M. P.; validation, B. Z. and R. M.; formal analysis, T. S, S. C., M. Z, and M. H.; investigation, B. Z.; resources, B. Z.; data curation, T. S.; writing—original draft preparation, B. Z., and T. S.; writing—review and editing, B. Z. and B. S.; visualization, M. Z.; supervision, R. M.; project administration, B. Z. All the authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
All the procedures performed in the study involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The study was approved by the Bioethics Committee of the Jagiellonian University (122.6120.94.2016).
Informed Consent
Informed consent was obtained from all the subjects involved in the study.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zapała, B., Stefura, T., Piwowar, M. et al. The Role of Single Nucleotide Polymorphisms of Monoamine Oxidase B, Dopamine D2 Receptor, and DOPA Decarboxylase Receptors Among Patients Treated for Parkinson’s Disease. J Mol Neurosci 72, 812–819 (2022). https://doi.org/10.1007/s12031-022-01966-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-022-01966-3