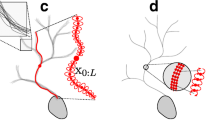
Abstract
The digital reconstruction of single neurons from 3D confocal microscopic images is an important tool for understanding the neuron morphology and function. However the accurate automatic neuron reconstruction remains a challenging task due to the varying image quality and the complexity in the neuronal arborisation. Targeting the common challenges of neuron tracing, we propose a novel automatic 3D neuron reconstruction algorithm, named Rivulet, which is based on the multi-stencils fast-marching and iterative back-tracking. The proposed Rivulet algorithm is capable of tracing discontinuous areas without being interrupted by densely distributed noises. By evaluating the proposed pipeline with the data provided by the Diadem challenge and the recent BigNeuron project, Rivulet is shown to be robust to challenging microscopic imagestacks. We discussed the algorithm design in technical details regarding the relationships between the proposed algorithm and the other state-of-the-art neuron tracing algorithms.














Similar content being viewed by others
References
Adalsteinsson, D, & Sethian, JA (1995). A fast level set method for propagating interfaces. Journal of Computational Physics, 118(2), 269–277.
Alexander, AL, Lee, JE, Lazar, M, & Field, AS (2007). Diffusion tensor imaging of the brain. Neurotherapeutics, 4(3), 316–329.
Basu, S, & Racoceanu, D (2014). Reconstructing neuronal morphology from microscopy stacks using fast marching. In 2014 IEEE international conference on image processing (ICIP) (pp 3597–3601).
Brown, KM, Barrionuevo, G, Canty, AJ, De Paola, V, Hirsch, JA, Jefferis, GS, Lu, J, Snippe, M, Sugihara, I, & Ascoli, GA (2011). The DIADEM data sets: representative light microscopy images of neuronal morphology to advance automation of digital reconstructions. Neuroinformatics, 9(2–3), 143–157.
Cesar, R Jr, & Costa, L (1999). Computer-vision-based extraction of neural dendrograms. Journal of Neuroscience Methods, 93(2), 121–131.
Chen, H, Xiao, H, Liu, T, & Peng, H (2015). Smarttracing: self-learning-based neuron reconstruction. Brain Informatics, 2(3), 135–144.
Feng, L, Zhao, T, & Kim, J (2015). NeuTube 1.0: a new design for efficient neuron reconstruction software based on the swc format. eNeuro, 2(1), ENEURO–0049.
Frangi, AF, Niessen, WJ, Vincken, KL, & Viergever, MA (1998). Multiscale vessel enhancement filtering. In Medical image computing and computer-assisted interventation (MICCAI) (pp. 130–137). Springer.
González, G, Türetken, E, Fleuret, F, & Fua, P (2010). Delineating trees in noisy 2D images and 3D image-stacks. In 2010 IEEE conference on computer vision and pattern recognition (CVPR) (pp. 2799–2806). IEEE.
Hassouna, MS, & Farag, AA (2007). Multistencils fast marching methods: a highly accurate solution to the eikonal equation on cartesian domains. IEEE Transactions on Pattern Analysis and Machine Intelligence, 29(9), 1563–1574.
Jameson, A, Schmidt, W, & Turkel, E (1981). Numerical solutions of the euler equations by finite volume methods using runge-kutta time-stepping schemes. AIAA paper 1259:1981.
Krissian, K, Malandain, G, Ayache, N, Vaillant, R, & Trousset, Y (2000). Model-based detection of tubular structures in 3D images. Computer Vision and Image Understanding, 80(2), 130–171.
Leandro, J, Cesar, R Jr, & Costa, LF (2009). Automatic contour extraction from 2D neuron images. Journal of Neuroscience Methods, 177(2), 497–509.
Long, F, Zhou, J, & Peng, H (2012). Visualization and analysis of 3D microscopic images. PLoS Computational Biology, 8(6), e1002519–e1002519.
Ming, X, Li, A, Wu, J, Yan, C, Ding, W, Gong, H, Zeng, S, & Liu, Q (2013). Rapid reconstruction of 3D neuronal morphology from light microscopy images with augmented rayburst sampling. PLoS ONE, 8(12), e84557.
Mukherjee, A, & Stepanyants, A (2012). Automated reconstruction of neural trees using front re-initialization. In SPIE medical imaging, International society for optics and photonics (pp. 83,141I–83,141I).
Mukherjee, S, Condron, B, & Acton, ST (2015). Tubularity flow field—a technique for automatic neuron segmentation. IEEE Transactions on Image Processing, 24(1), 374–389.
Parekh, R, & Ascoli, GA (2013). Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron, 77(6), 1017–1038.
Pawley, JB. (2006). Handbook of biological confocal microscopy, (pp. 20–42). Boston: Springer US , chap Fundamental Limits in Confocal Microscopy.
Peng, H, Ruan, Z, Long, F, Simpson, JH, & Myers, EW (2010). V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nature Biotechnology, 28(4), 348–353.
Peng, H, Long, F, & Myers, G (2011). Automatic 3D neuron tracing using all-path pruning. Bioinformatics, 27(13), i239–i247.
Peng, H, Bria, A, Zhou, Z, Iannello, G, & Long, F (2014). Extensible visualization and analysis for multidimensional images using Vaa3D. Nature Protocols, 9(1), 193–208.
Peng, H, Hawrylycz, M, Roskams, J, Hill, S, Spruston, N, Meijering, E, & Ascoli, G (2015a). BigNeuron: large-scale 3D neuron reconstruction from optical microscopy images. Neuron, 87(2), 252–256.
Peng, H, Meijering, E, & Ascoli, G (2015b). From DIADEM to BigNeuron. Neuroinformatics, 13(3), 259–260.
Santamaría-Pang, A, Hernandez-Herrera, P, Papadakis, M, Saggau, P, & Kakadiaris, IA (2015). Automatic morphological reconstruction of neurons from multiphoton and confocal microscopy images using 3D tubular models. Neuroinformatics, 13(3), 297–320.
Sethian, JA. (1999). Level set methods and fast marching methods: evolving interfaces in computational geometry, fluid mechanics, computer vision, and materials science (Vol. 3). Cambridge: Cambridge University Press.
Tsitsiklis, JN (1995). Efficient algorithms for globally optimal trajectories. IEEE Transactions on Automatic Control, 40(9), 1528–1538.
Türetken, E, González, G, Blum, C, & Fua, P (2011). Automated reconstruction of dendritic and axonal trees by global optimization with geometric priors. Neuroinformatics, 9(2–3), 279– 302.
Van Uitert, R, & Bitter, I (2007). Subvoxel precise skeletons of volumetric data based on fast marching methods. Medical Physics, 34(2), 627–638.
Wang, Y, Narayanaswamy, A, Tsai, CL, & Roysam, B (2011). A broadly applicable 3-D neuron tracing method based on open-curve snake. Neuroinformatics, 9(2–3), 193–217.
Wearne, S, Rodriguez, A, Ehlenberger, D, Rocher, A, Henderson, S, & Hof, P (2005). New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience, 136 (3), 661–680.
Xiao, H, & Peng, H (2013). APP2: automatic tracing of 3D neuron morphology based on hierarchical pruning of a gray-weighted image distance-tree. Bioinformatics, 29(11), 1448– 1454.
Yang, J, Gonzalez-Bellido, PT, & Peng, H (2013). A distance-field based automatic neuron tracing method. BMC Bioinformatics, 14(1), 93.
Yuan, X, Trachtenberg, JT, Potter, SM, & Roysam, B (2009). MDL constrained 3D grayscale skeletonization algorithm for automated extraction of dendrites and spines from fluorescence confocal images. Neuroinformatics, 7 (4), 213–232.
Zhang, D, Liu, S, Liu, S, Feng, D, Peng, H, & Cai, W (2016). Reconstruction of 3D neuron morphology using Rivulet back-tracking. In The IEEE international symposium on biomedical imaging: from nano to macro. IEEE.
Zhao, T, Xie, J, Amat, F, Clack, N, Ahammad, P, Peng, H, Long, F, & Myers, E (2011). Automated reconstruction of neuronal morphology based on local geometrical and global structural models. Neuroinformatics, 9(2–3), 247–261.
Zhou, Z, Sorensen, S, Zeng, H, Hawrylycz, M, & Peng, H (2014). Adaptive image enhancement for tracing 3D morphologies of neurons and brain vasculatures. Neuroinformatics, 13(2), 153–166.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Liu, S., Zhang, D., Liu, S. et al. Rivulet: 3D Neuron Morphology Tracing with Iterative Back-Tracking. Neuroinform 14, 387–401 (2016). https://doi.org/10.1007/s12021-016-9302-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-016-9302-0